We conducted a comprehensive literature search through Cochrane, Ovid, and PubMed databases, focusing on studies published from the beginning of January 2000 to July 5, 2022. Our analysis indicated a reduction in the gene expressions of CXCR1 and TLR4, while there was an upregulation of CXCR2, TRIF, and SIGIRR in rUTI patients as opposed to healthy subjects. The most critical pathways were TLR signaling, I-kappa B kinase/NF-kappa B signaling, and MyD88-independent TLR signaling pathways.
In conclusion, our review explored the variability in gene expression among several substantial genes and highlighted numerous biological pathways implicated in rUTIs. This research begins to lay the groundwork for unlocking the hidden underlying mechanisms involved in recurrent UTIs, paving the way for innovative therapeutic and preventive measures. The elucidation of these genetic factors might also aid in designing personalized treatments. Additionally, these findings could be a foundation for future research in developing targeted gene therapies or biomarkers for recurrent UTIs.
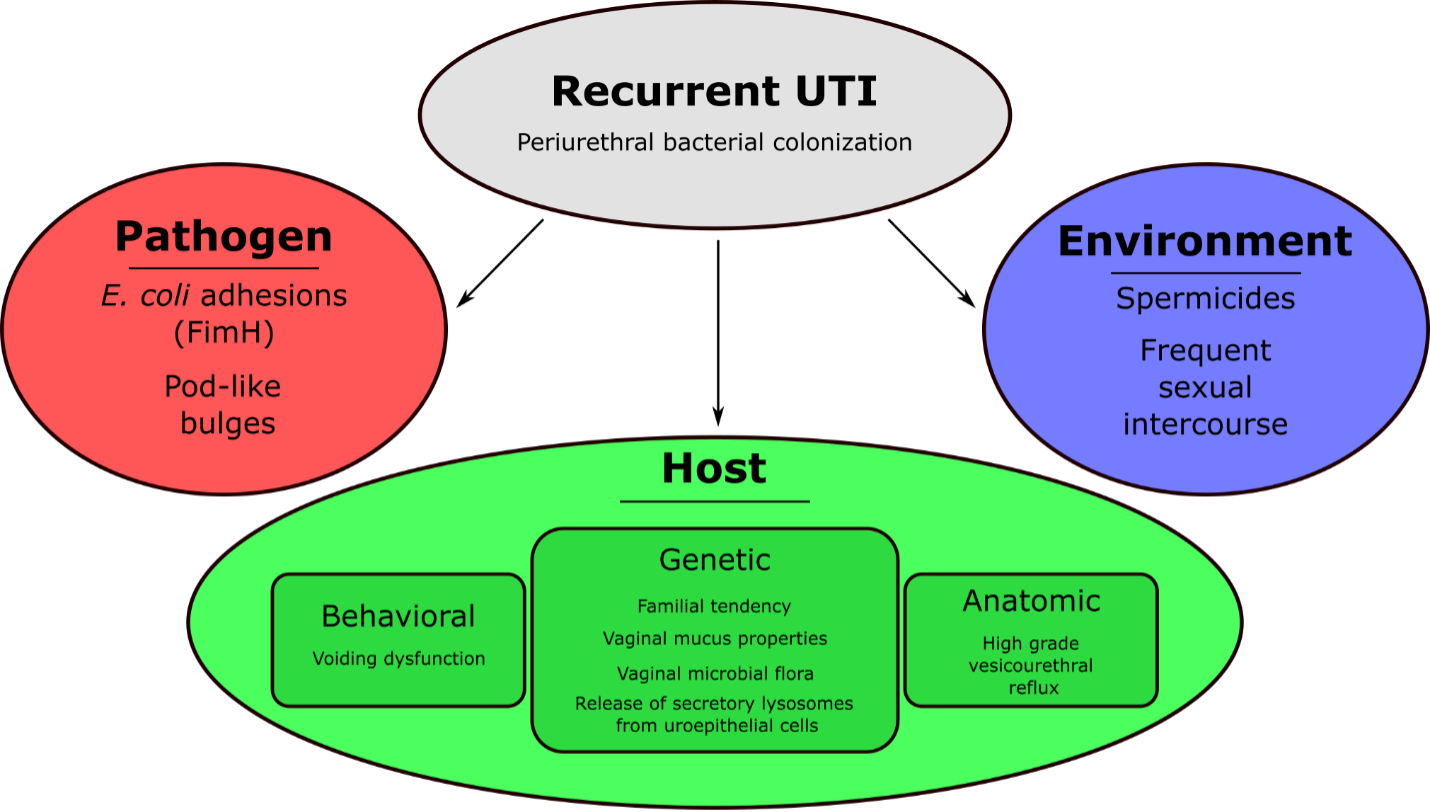
Figure 1. was adapted from Isali et al. Multifaceted elements involved in the emergence of recurrent UTIs.2
Written by: Ilaha Isali, MD, Thomas R. Wong, Adonis Hijaz, MD, & David Sheyn, MD
Department of Urology, University Hospitals, Cleveland Medical Center, Cleveland, OH
References:
- M. Zaffanello, G. Malerba, L. Cataldi, F. Antoniazzi, M. Franchini, E. Monti, V. Fanos, Genetic risk for recurrent urinary tract infections in humans: a systematic review, J Biomed Biotechnol 2010 (2010) 321082.
- I. Isali, T.R. Wong, A.F. Batur, C.W. Wu, F.R. Schumacher, R. Pope, A. Hijaz, D. Sheyn, Recurrent urinary tract infection genetic risk: a systematic review and gene network analysis, Int Urogynecol J (2023).


