(UroToday.com) The 2024 Advanced Prostate Cancer Consensus Conference (APCCC) meeting featured a session on the management of metastatic CRPC (mCRPC), and a presentation by Dr. Niven Mehra discussing when we should do tumor genomic profiling in advanced prostate cancer. Dr. Mehra started by noting that the EAU, ESMO, NCCN, and APCCC governing bodies provide guidance regarding who to test, when to test, and the genes to test:

Strategies for testing and re-testing are as summarized by Dr. Mehra:
- Metachronous: archived (random/targeted prostate biopsies, RALP specimens, lymph node tissue from pelvic lymph node dissections) or fresh biopsies
- Synchronous: prostate biopsy, metastatic biopsy, or liquid biopsy
- mCRPC: archived, fresh biopsy, or liquid biopsy
The latest update of the NCCN guidelines suggest that for somatic tumor testing “the tumor molecular profiles may change with subsequent treatments and re-evaluation may be considered at the time of cancer progression for treatment decision-making.” Dr. Mehra then provided his personal interpretation of the guidelines and concurrent NGS testing strategy, which is influenced by a lack of resources for repeat testing:
- mHSPC: infrequent – high risk mutations include TP53, PTEN, RB1, BRCA2, ATM
- mCRPC: when it may influence treatment decisions – first-line mCRPC treatment, BRCA1, BRCA2, non-BRCA HRR, HRD, MMRd/MSI-H, high TMB, PTEN, RB1, TP53, AR, AKT, and PIK3CA
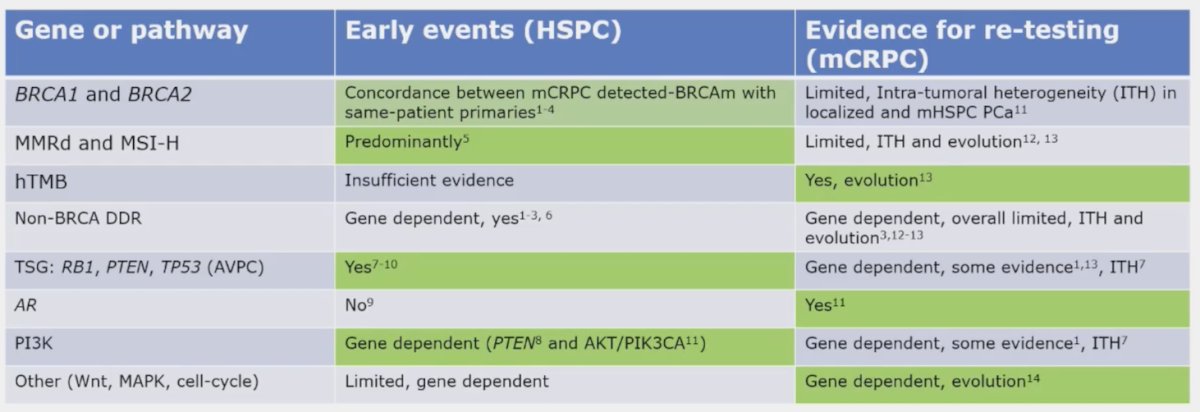
The cancer biology and genomic landscape varies across localized and the metastatic disease state, as well as for mHSPC and mCRPC:

When considering the use of diagnostic prostate cancer biopsies for NGS, be aware of intra-tumor heterogeneity. There can be analytical or technical reasons for mis-detection and mis-interpretation, including the use of diagnostic biopsies/under sampling of a heterogeneous primary that does not accurately capture intra-tumor heterogeneity:

With regards to the use of liquid biopsies for NGS testing, there are also analytical or technical reasons for mis-detection and mis-interpretation, such as low ctDNA fraction leading to false negatives:
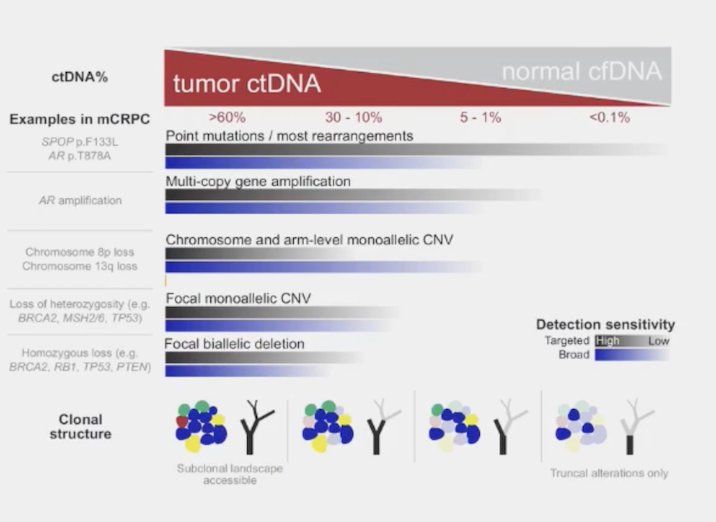
Of note, the median ctDNA percentage is between 0-18%, constraining comprehensive genomic analysis in up to ~50% of samples:

Moreover, the use of liquid biopsies without correction with WBC DNA leads to false positives secondary to clonal hematopoiesis of indeterminate potential (CHiP). Tumor fraction also constrains the alterations that can be identified, given that a median tumor fraction percentage >40% in mCRPC with a median ctDNA percentage <205 missing complex SV, in particular homozygous deletions. For what to test on which tissue in metastatic prostate cancer, Dr. Mehra provides the following table to display genes and context for testing:
The benefit of genomic re-testing in advanced prostate cancer is longitudinal sampling that may identify new actionable events in more advanced disease settings. In a multicenter retrospective analysis of 1,597 patients in the Prostate Cancer Precision Medicine Multi-Institutional Collaborative Effort (PROMISE) database used for characterizing patterns of single versus serial NGS testing, Park et al.1 identified the following actionable alterations: BRCA1, BRCA2, ATM, BRIP1, BARD1, CDK12, CHEK1, CHEK2, FANCL, PALB2, PPP2R2A, RAD51B, RAD51C, RAD51D, RAD4L, MSH2, MSH3, MSH6, MLH1, MLH3, PMS1, PMS2, MSI-H, and high TMB >= 10 mut/MB. As follows are the mutations detected stratified by number of NGS tests:
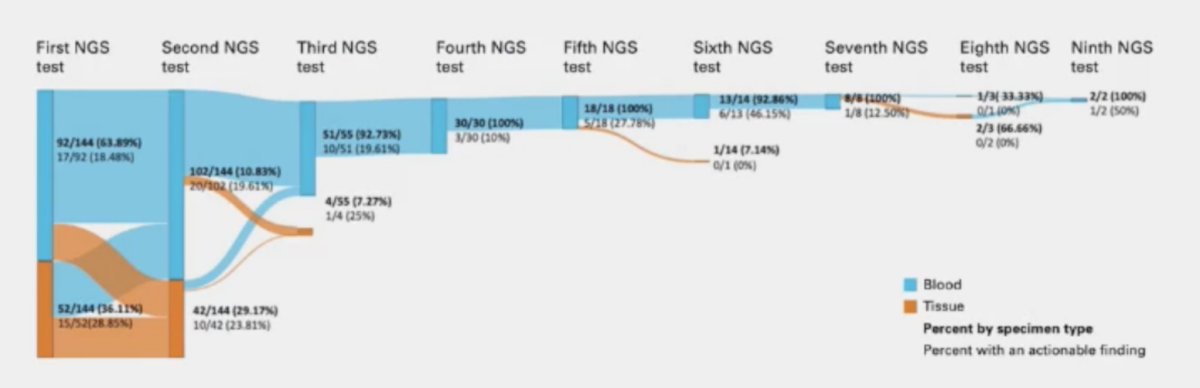
New actionable data were found on 11.1% (16 of 144) of second NGS tests for the serial group. Cases detected on or beyond second testing included:
- BRCA2: 30% (3 of 10)
- Non-BRCA HRR genes: 55% (16 of 29)
- MSI-H: 33% (2 of 6)
- TMB high: 92% (9 of 11)
Finally, Dr. Mehra discussed the prospective ctDNA ProBIO trial assessing genomic re-testing in advanced prostate cancer:

Dr. Mehra concluded his presentation by discussing when we should do tumor genomic profiling in advanced prostate cancer by emphasizing that tissue is the issue for testing and re-testing:
- Prostate primary tumor content allows for 50-90% ‘successful’ NGS reporting in HSPC or CPRC. When NGS reports with inferred tumor fraction >30%, there is a low probability of new alterations on re-testing:
- BRCA1/BRCA2
- Canonical MMRd
- PI3k pathway
- Unsuccessful or incomplete NGS reporting, ie inferred tumor fraction percentage is below 20%, we should consider repeat testing for homozygous TSG deletion and other CNV:
- Fresh tissue biopsy
- Liquid with ctDNA percentage preferably >20%
- Repeat testing in mCRPC? Do this for late state (ie. visceral metastases or NEPC)
- Fresh tissue biopsy
- Liquid: possibly MSI/H, high TMB, non-BRCA HRR, Wnt, MAPK, and cell cycle rearrangements
Presented by: Niven Mehra, MD, PhD, Radboud University Medical Center, Nijmegen, The Netherlands
Written by: Zachary Klaassen, MD, MSc – Urologic Oncologist, Associate Professor of Urology, Georgia Cancer Center, Wellstar MCG Health, @zklaassen_md on Twitter during the 2024 Advanced Prostate Cancer Consensus Conference (APCCC) Meeting, Lugano, Switzerland, Thurs, Apr 25 - Sat, Apr 27, 2024.
References:


