Chicago, IL (UroToday.com) Although enzalutamide prolongs life in metastatic castration-resistant prostate cancer (mCRPC) patients, the development of drug resistance and subsequent disease progression is nearly universal. Several enzalutamide resistance mechanisms have been described in pre-clinical models, however, there are limited analyses examining acquired resistance mechanisms in tissue biopsies from patients treated with enzalutamide.
Genomic sequencing efforts have identified several recurrent genomic alterations in specific genes, including AR, TP53, PTEN, and ETS fusions, however genomic lesions enriched in enzalutamide resistant tumor samples are not well characterized. Dr. Xiangnan Guan and colleagues presented results of their study analyzing whole-genome sequencing and RNA sequencing of tumors obtained from patients with enzalutamide-naive or enzalutamide-resistant mCRPC.
In this study, 101 men with mCRPC who underwent image-guided biopsy and subsequent whole genome sequencing were included (n = 64 with enzalutamide -naive and n = 37 with enzalutamide -resistant mCRPC). The differential copy number alteration (CNA) events enriched in enzalutamide-resistant vs. naïve samples were determined, and the prognostic significance of differential CNAs was assessed. RNA-seq data were evaluated to confirm that CNAs correlated with changes in gene expression of relevant loci and to identify potentially druggable targets selectively activated in tumors with specific CNAs.
Copy number loss was more common than gain in enzalutamide-resistant tumors. Specifically, the authors identified 123 protein-coding genes that were more commonly lost in enzalutamide-resistant samples, eight of which were previously described tumor suppressor genes:
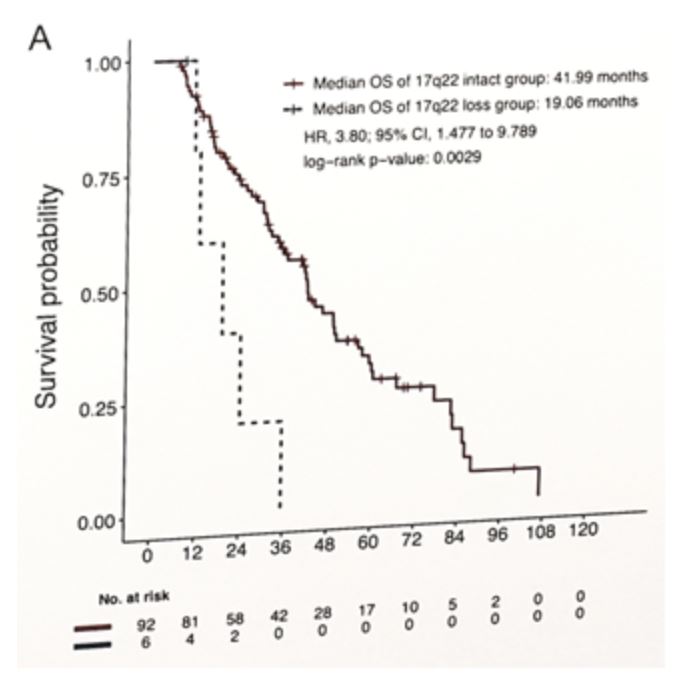
There was a strong concordance of copy number loss and reduced mRNA expression of these genes. Dr. Guan’s team then identified one gene (17q22 containing RNF43 and SRSF1) from this list of eight genes whose copy number loss was associated with poor overall survival; median OS from date of CRPC was 19.1 months in tumors with gene loss vs. 42.0 months in intact tumors, HR 3.80, 95% CI 1.48–9.49:
Master Regulator analysis determined that tumors with copy number loss of poor prognosis 17q22 had activation of several potentially targetable factors, including the kinases Akt, CDK1/2, and PLK1.
Dr. Guan’s conclusions were as follows:
-
The frequency of previously known prostate cancer tumor suppressor genes including PTEN, RB1, and TP53 does not differ between enzalutamide-naïve and enzalutamide-resistant tumors
-
Copy number loss is common in enzalutamide resistance tumors and these loci contain several putative tumor suppressor genes
-
Focal deletion of chromosome 17q22 defines a previously unappreciated molecular subset of enzalutamide-resistant CRPC associated with poor clinical outcome
-
There are several potential druggable targets in 17q22 loss, including Akt1, CDK1/2, and PLK1
Presented by: Xiangnan Guan, PhD, Knight Cancer Institute, Oregon Health & Science University, Portland, OR
Written by: Zachary Klaassen, MD, MSc, Assistant Professor of Urology, Georgia Cancer Center, Augusta University/Medical College of Georgia, @zklaassen_md at the 2019 ASCO Annual Meeting #ASCO19, May 31- June 4, 2019, Chicago, IL USA