(UroToday.com) The 2024 American Society of Clinical Oncology (ASCO) Annual Meeting held in Chicago, IL was host to a kidney and bladder rapid oral abstract session. Dr. Soki Kashima presented the results of a single-cell analysis of renal cell carcinoma (RCC) investigating T cell phenotypes associated with response or resistance to immune checkpoint inhibitors (ICIs).
While immuno-oncology (IO)-based combination therapies have emerged as the recommended 1st line treatment approach for patients with advanced RCC, >70% of patients ultimately experience disease progression. As such, further evaluation of the immunologic underpinnings driving treatment response/resistance is needed.
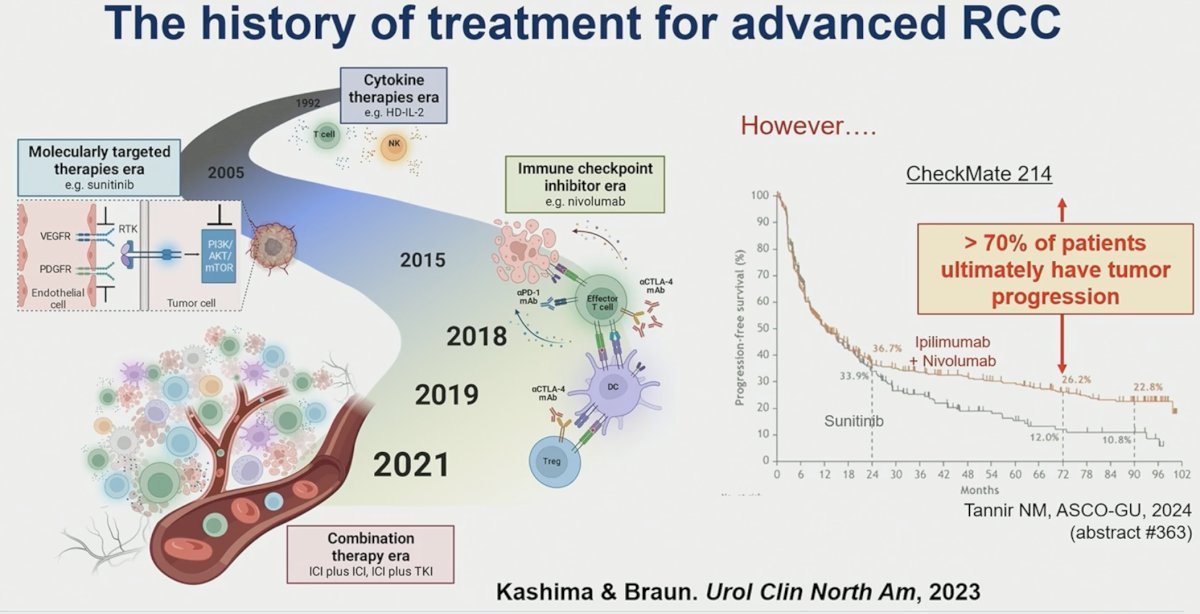
In this study, Dr. Kashima and colleagues performed a comprehensive characterization of the tumor microenvironment to elucidate mechanisms of response/resistance to immune checkpoint inhibitors (ICI) in RCC.
A total of 70 tumor samples (clear cell, n=59; non-clear cell, n=11) from 63 patients with RCC were collected before (n = 48) and/or after (n = 22) systemic therapies (VEGFi, n = 9; ICI monotherapy, n = 20; ICI + ICI, n = 17; ICI + VEGFi, n = 9; others, n = 15). This cohort contained 12 paired samples on pre and post from five patients, and 58 unpaired samples. Responders were those that experienced either complete or partial responses (n = 22), and non-responders (NR) were those who experienced disease progression (n = 33), per RECIST criteria. They performed single-cell RNA-sequence (scRNA-seq) on all samples and established a transcriptomics atlas in RCC. They utilized established gene expression signatures to interrogate cellular composition and functional states for samples from ICI-treated patients. They used non-negative matrix factorization (NMF) to identify gene programs, offering superior feature preservation and interpretability. 443,337 high-quality viable cells were annotated to lymphoid, myeloid, tumor, endothelial, or fibroblast compartments, capturing the RCC tumor microenvironment landscape.

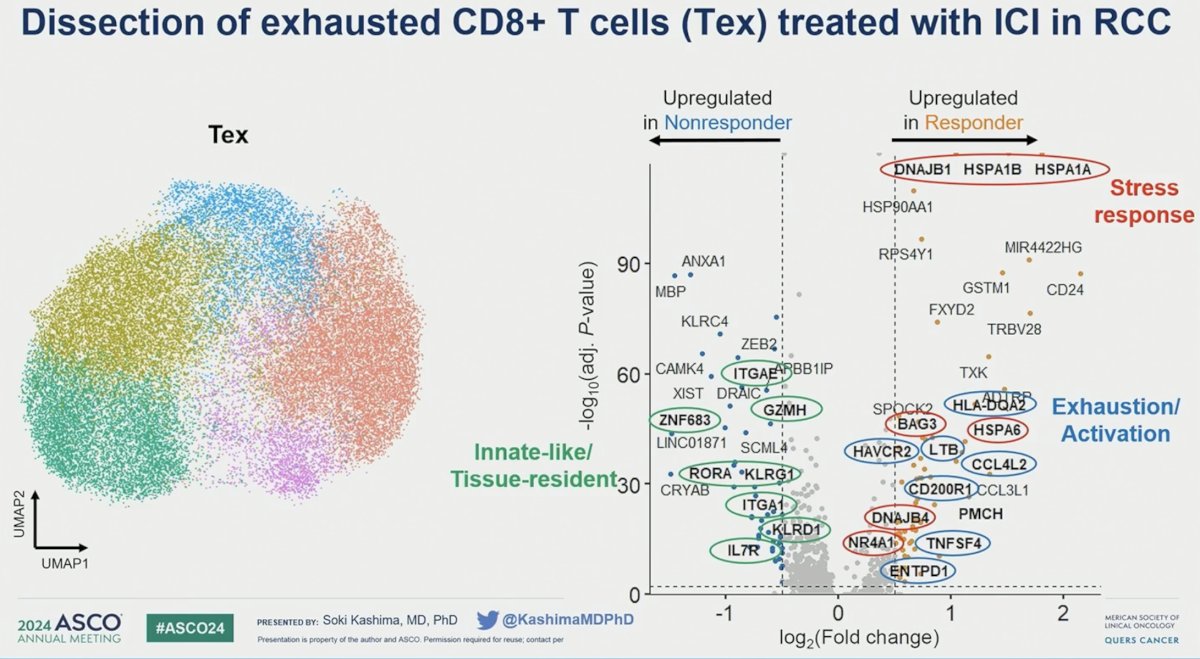
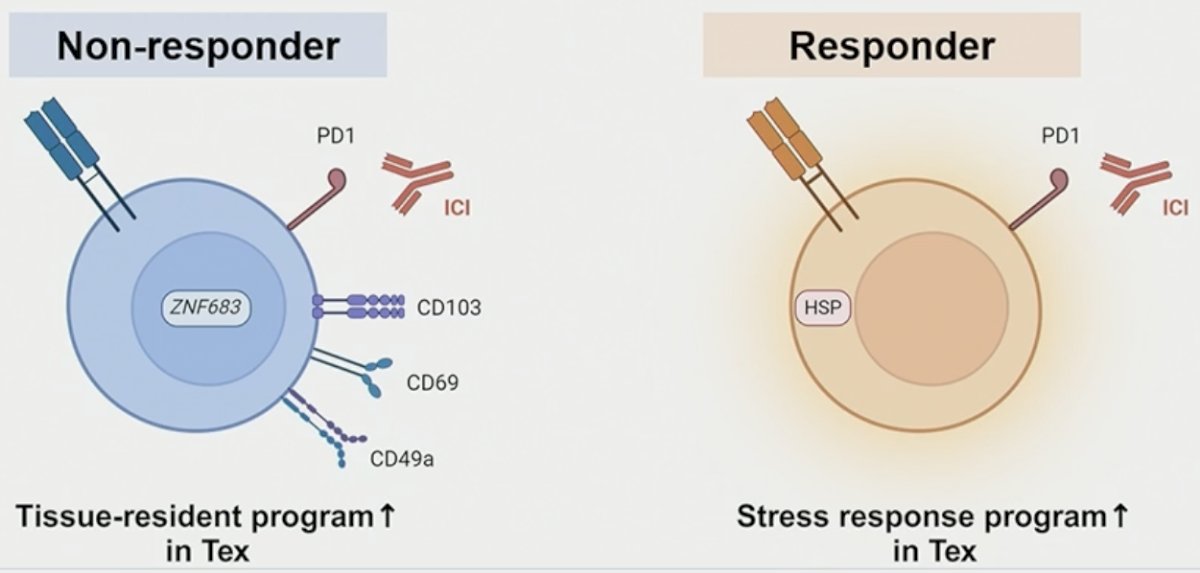
Among CD8+ T cells, the investigators observed significant heterogeneity, particularly in exhausted T cells (Tex) expressing PD-1 and TIM-3. Exhausted T-cells in non-responders demonstrated enrichment for tissue-residency and innate-like genes and gene programs, exemplified by significant upregulation of ZNF683 (p=0.031) and ITGAE (p=0.0041).

In contrast, exhausted T-cells in responders exhibited a marked upregulation of heat shock protein genes, such as HSP1B (p < 0.001) and DNAJB1 (p < .001), highlighting a distinct genomic profile.
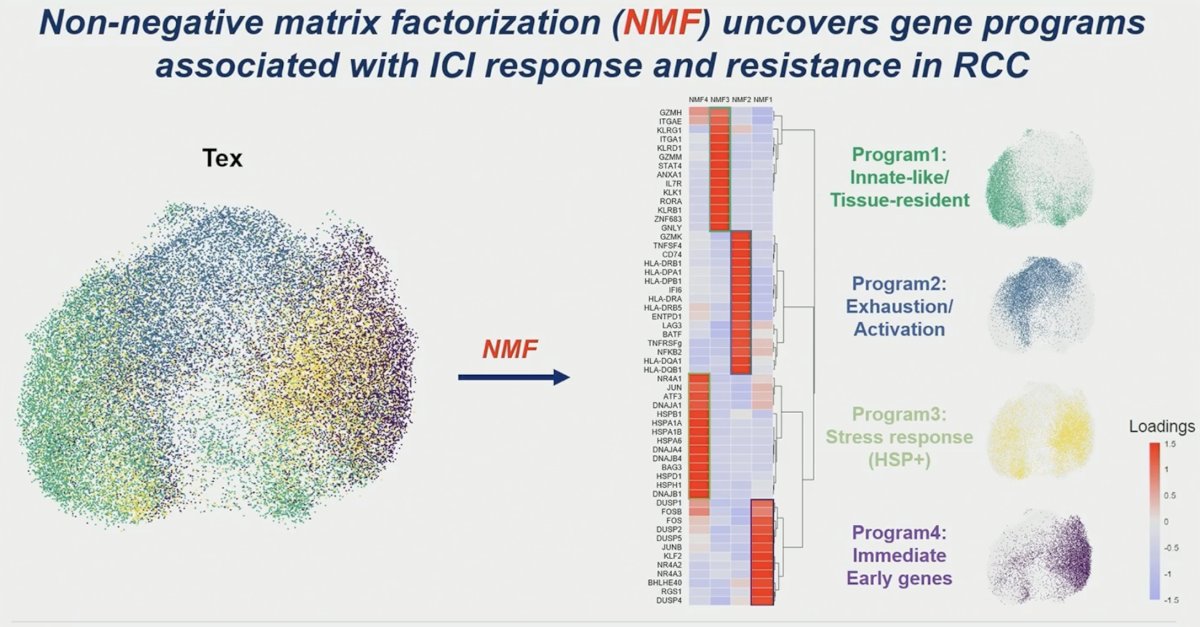
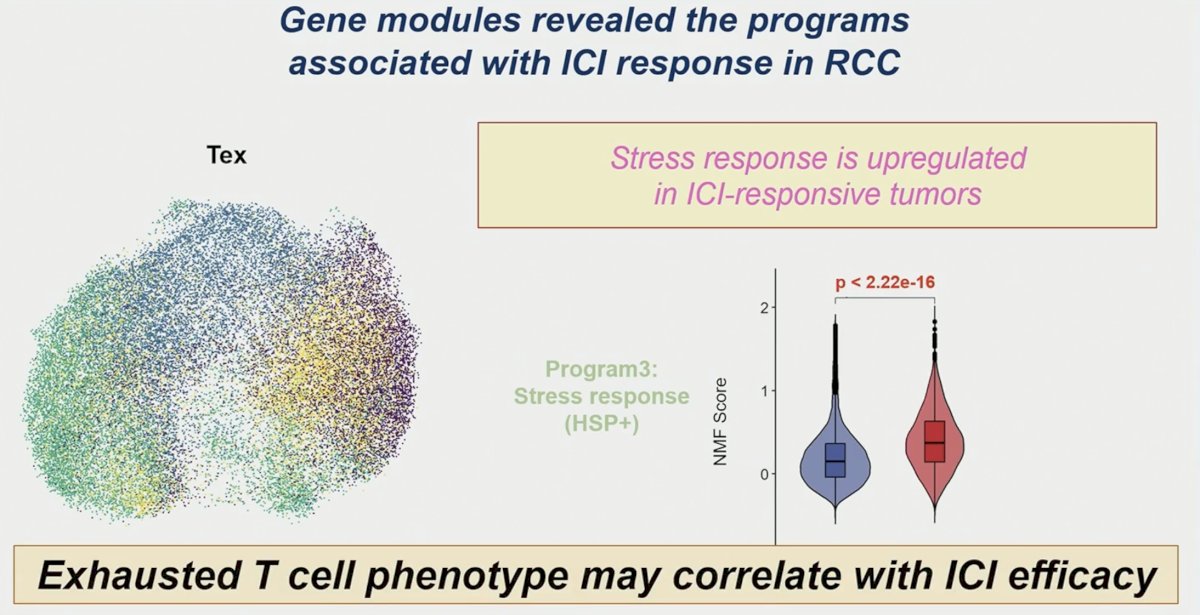
Notably, through NMF analysis, exhausted T-cells in responders showed a significantly higher stress response program and terminal exhaustion program than in non-responders at baseline and after ICI treatment.
Further analysis through gene signature scoring showed an association between exhausted T-cells in responders and enhanced IFN and chemokine activities, stress response, and terminal differentiation post-ICI.
Dr. Kashima concluded as follows:
- In exhausted CD8 T cells:
- Tissue-resident program is associated with ICI resistance.
- Stress response program is associated with ICI response.
- ScRNA-seq as a powerful strategy for deep biological analysis in large cohorts
Presented by: Soki Kashima, MD, PhD, Postdoctoral Associate at the Braun Lab, Yale Cancer Center, Yale School of Medicine, New Haven, CT
Written by: Rashid Sayyid, MD, MSc – Society of Urologic Oncology (SUO) Clinical Fellow at The University of Toronto, @rksayyid on Twitter during the 2024 American Society of Clinical Oncology (ASCO) Annual Meeting, Chicago, IL, Fri, May 31 – Tues, June 4, 2024.


