(UroToday.com) The 2024 American Society of Clinical Oncology (ASCO) Annual Meeting held in Chicago, IL was host to a kidney and bladder cancers poster session. Dr. Rana McKay presented the results of a multimodal real-world analysis of matched tissue and circulating tumor DNA (ctDNA) in renal cell carcinoma (RCC) patients.
Next generation sequencing (NGS) of ctDNA can complement tissue NGS and is a non-invasive test that can be conducted serially, with the potential to enhance assessment of spatial and temporal molecular tumor heterogeneity. In this study, Dr. McKay and colleagues investigated mutations in RCC patients from matched tissue and ctDNA genomic profiling.
Using the Tempus multimodal database, the investigators retrospectively analyzed de-identified NGS data from RCC patients that had both tissue (Tempus xT, 648 genes) and ctDNA testing (Tempus xF, 105 genes) performed. They included patients with matched samples (collected +/- 90 days of one another). Clinical characteristics and select pathogenic somatic short variants and copy number variants (amplifications and deletions, two copy number losses) were evaluated. Concordance analyses were restricted to the 105 genes tested on the ctDNA panel and further restricted to short variants, with the exception of amplifications and copy number losses detected by both Tempus xF and xT.
The study included 393 patients. The baseline patient characteristics are summarized below. The median patient age was 61 years and 71% were male. The median time from tissue to blood collection was 21 days (IQR: 7 –39 days).

Metastatic disease was present at the time of tissue and blood collection in 67% and 68% of patients, respectively. The most common sites of tissue were:
- Kidney (49%, n=189)
- Bone (11%, n=43)
- Lung (9%, n=34)
- Lymph node (8%, n=29)
- Liver (6%, n=23)
- Brain/CNS (4%, n=17)

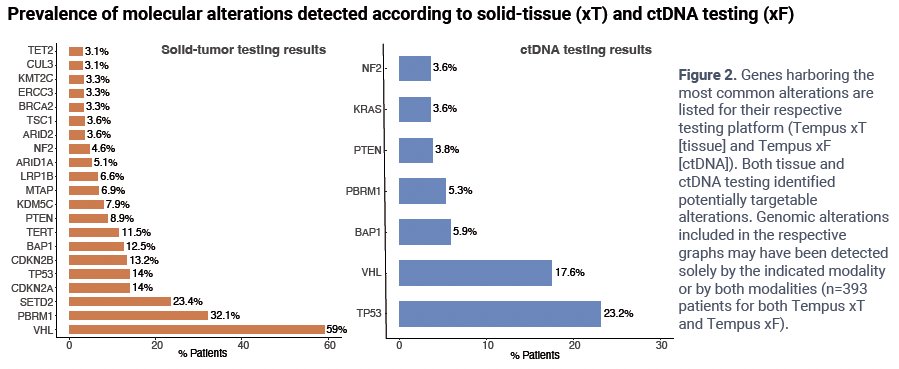
Genes harboring the most common pathogenic somatic short variants in tissue included VHL (59%, n=232), PBRM1 (31%, n=123), SETD2 (23%, n=91), and TP53 (14%, n=54). Genes with common pathogenic somatic short variants in ctDNA included TP53 (23%, n=91), VHL (18%, n=69), BAP1 (6%, n=23), and PBRM1 (5%, n=21).
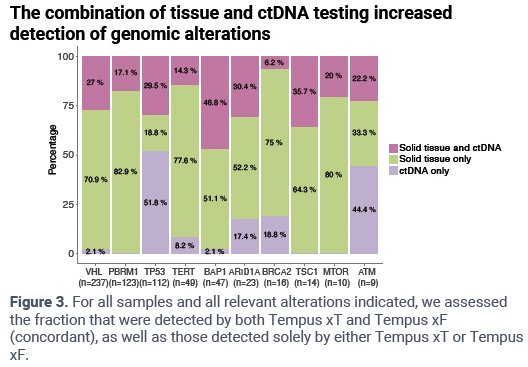
The combination of tissue and ctDNA testing increased detection of mutations.
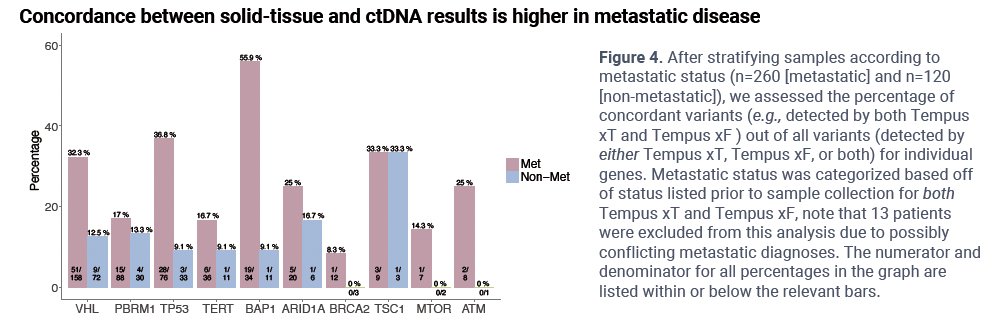
There was higher concordance between somatic alterations in select genes among patients with metastases.
Dr. McKay concluded as follows:
- ctDNA profiling is complementary to tissue-based NGS in RCC and can increase the detection of genomic alterations.
- Concordance between ctDNA and tissue profiling was higher in individuals with metastatic disease.
- Future research is warranted to understand how longitudinal ctDNA analysis can define biomarkers of response and resistance at the mutation and ctDNA fraction levels.
Presented by: Rana McKay, MD, Associate Professor, Department of Medicine, University of California, San Diego, CA
Written by: Rashid Sayyid, MD, MSc – Society of Urologic Oncology (SUO) Clinical Fellow at The University of Toronto, @rksayyid on Twitter during the 2024 American Society of Clinical Oncology (ASCO) Annual Meeting, Chicago, IL, Fri, May 31 – Tues, June 4, 2024.


