(UroToday.com) The 2024 American Society of Clinical Oncology (ASCO) Annual Meeting held in Chicago, IL between May 31st and June 4th, 2024 was host to a kidney and bladder cancers poster session. Dr. Niha Beig presented a machine learning-derived model using histologic features for the prediction of transcriptomic molecular subtypes in advanced renal cell carcinoma (RCC).
Metastatic RCC is a molecularly heterogeneous disease with varying levels of angiogenic presence, immune infiltration, and PD-L1 expression. Transcriptomic analysis in the phase 3 IMmotion151 trial identified 7 molecular subtypes that showed differential outcomes to atezolizumab + bevacizumab versus sunitinib treatment.1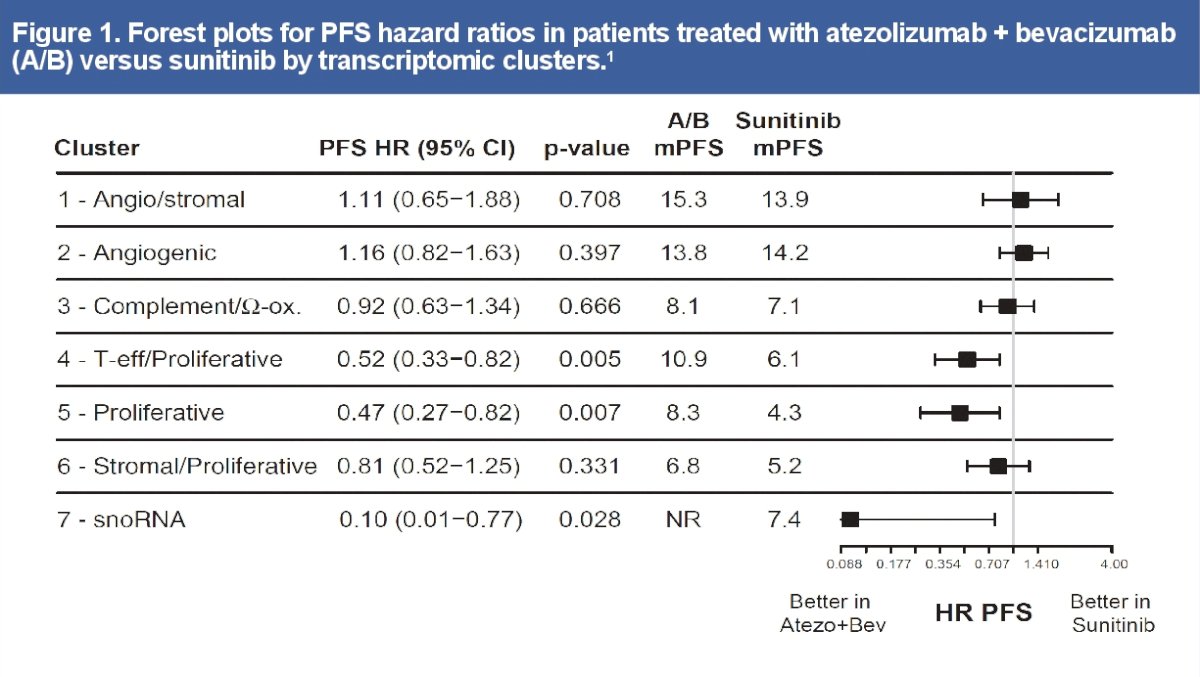
In this study, Dr. Beig and colleagues presented histological correlates of these molecular subtypes as identified in whole slide images of hematoxylin and eosin (H&E)-stained tumors. The study objective was to develop machine learning models to derive histological features in metastatic RCC tumors and to identify histological correlates of RCC molecular subtypes on H&E whole slide images and evaluate them as surrogate imaging-based predictive biomarkers.
This exploratory analysis using whole slide images evaluated imaging-based features and their association with molecular subtypes and clinical outcomes in untreated metastatic RCC. Machine learning models identified 922 H&E-derived human interpretable histological features in RCC associated with tumor and stromal (including blood vessels, immune cells, and fibroblasts) cell and tissue morphologies, and nucleus shape. These machine-learning human interpretable histological features were then extracted from whole slide images in two metastatic RCC trials: IMmotion151 (n=97, discovery cohort) and IMmotion150 (n=203, validation cohort).2,3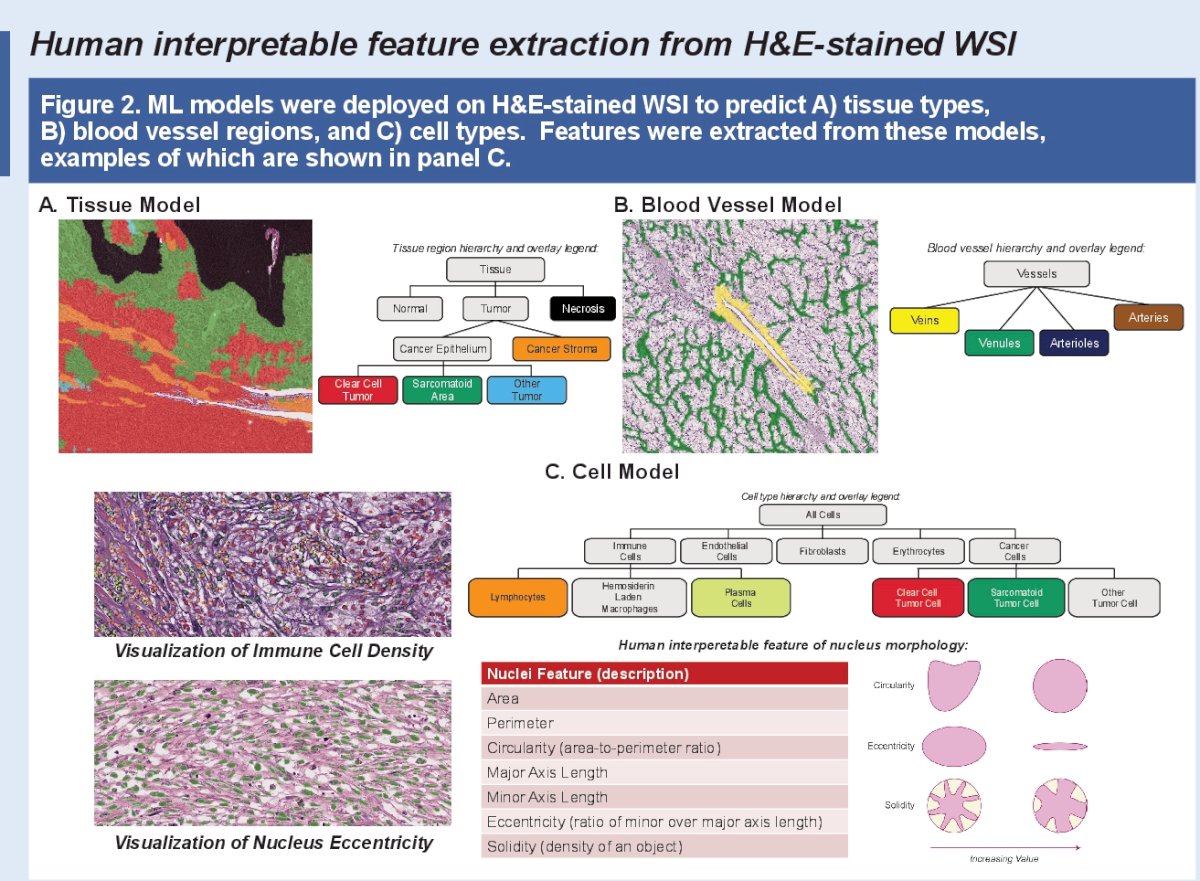
As previously described, seven molecular subtypes were combined into four subgroups for computational power purposes:
- Angiogenic (comprised of Angiogenic/Stromal and Angiogenic)
- Complement/Omega Oxidation
- T-effector
- Proliferative (comprised of Proliferative and Stromal Proliferative)
The snoRNA subset was excluded from this analysis due to a prohibitively low prevalence.
Univariate analysis with false discovery rate (FDR) correction was applied to identify positively associated human interpretable features in each of the 4 subgroups in the IMmotion 151 whole slide images and then validated in IMmotion150 molecular subgroups. Representative machine learning human interpretable features that showed uniquely higher abundance in each molecular subgroup in both studies were dichotomized by tertiles as 'high' or 'low/intermediate’ and were associated with progression-free survival to fit Cox proportional hazard models in the IMmotion 151 study.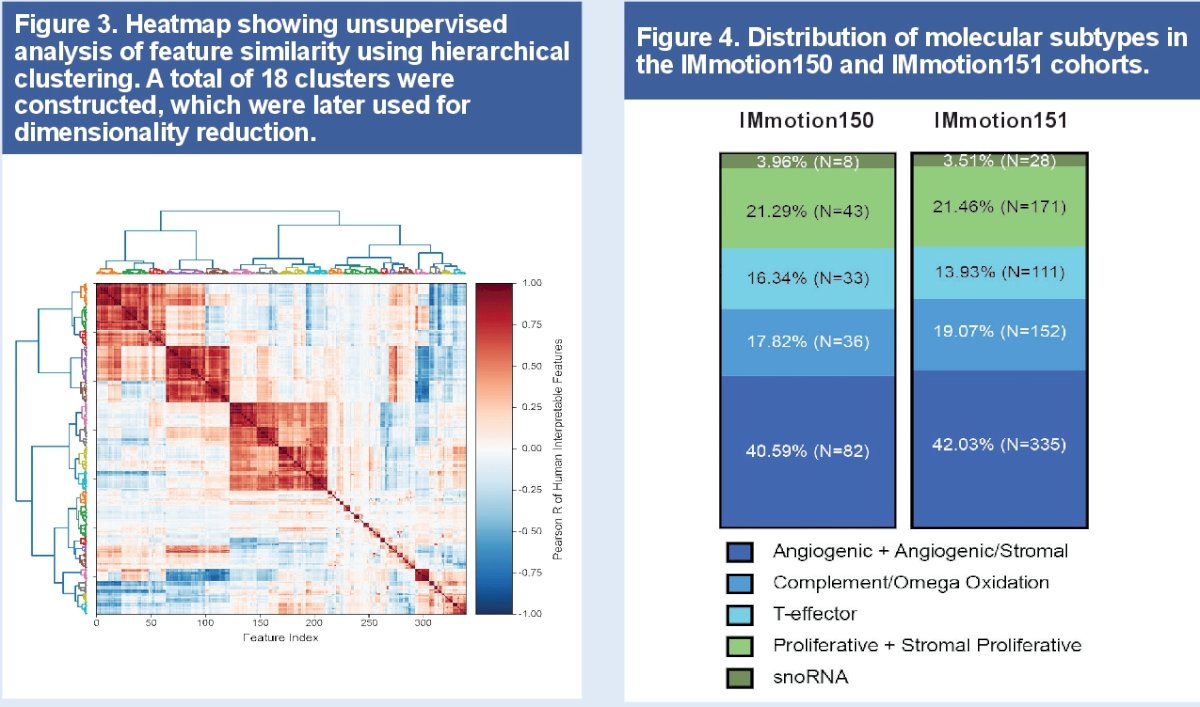
Distinct imaging-based phenotypes were associated with each RCC molecular subgroup:
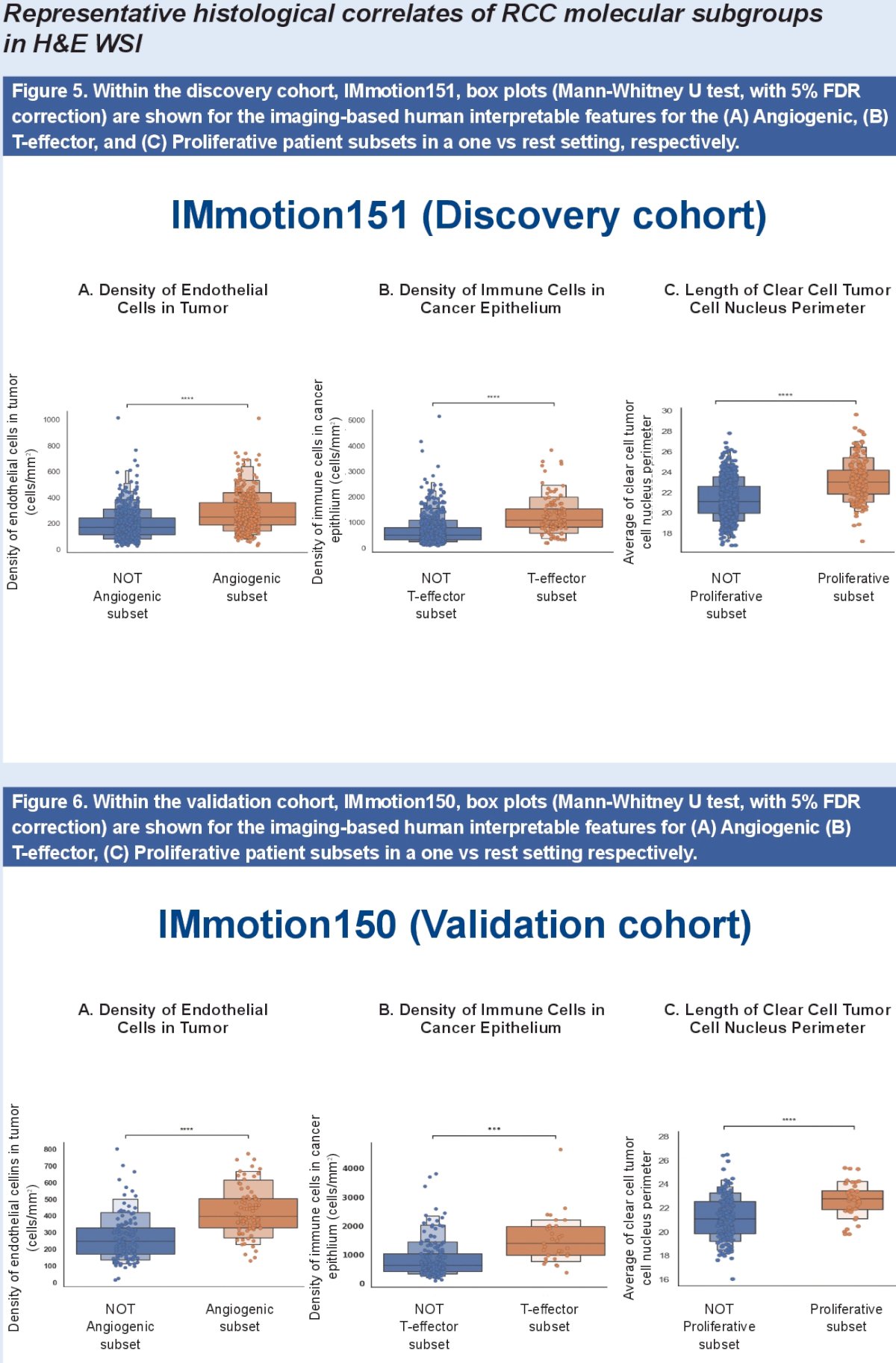
- The Angiogenic subgroup had a higher prevalence of 40 imaging-based features associated with density of endothelial cells and vessels in the cancer epithelium.
- The T-effector subtype showed higher abundance of 64 imaging-based features associated with immune cell presence in stroma.
- The Proliferative subgroup showed higher prevalence of 40 imaging-based features associated with nuclear morphologies.
- No imaging-based features were uniquely expressed for Complement/ Omega oxidation subgroup.
Representative human interpretable features enriched in T-effector and Proliferative subgroups showed improved PFS benefit with atezolizumab + bevacizumab, compared to sunitinib.
Dr. Beig concluded as follows:
- They identified unique histological features of RCC tumors that correlate with previously defined molecular subtypes and are associated with differential clinical outcomes.
- These results suggest that clinically relevant RCC subtypes can be extracted directly from H&E-stained whole slide images and may complement gene expression-based patient stratification and selection strategies.
- Further prospective validation of a possible biomarker-directed approach to 1st line RCC treatment is warranted.
Presented by: Niha Beig, PhD, Department of Biomedical Engineering, Case Western Reserve University, Cleveland, OH, USA.
Written by: Rashid Sayyid, MD, MSc – Society of Urologic Oncology (SUO) Clinical Fellow at The University of Toronto, @rksayyid on Twitter during the 2024 American Society of Clinical Oncology (ASCO) Annual Meeting, Chicago, IL, May 31st – June 4th, 2024
References:- Motzer RJ, Banchereau R, Hamidi H, et al. Molecular Subsets in Renal Cancer Determine Outcome to Checkpoint and Angiogenesis Blockade. Cancer Cell. 2020;38(6): 803-17.e4.
- McDermott DF, Huseni MA, Atkins MB, et al. Clinical activity and molecular correlates of response to atezolizumab alone or in combination with bevacizumab alone or in combination with bevacizumab versus sunitinib in renal cell carcinoma. Nat Med. 2018 Jun;24(6):749-757.
- Rini BI, Powles T, Atkins MB, et al. Atezolizumab plus bevacizumab versus sunitinib in patients with previously untreated metastatic renal cell carcinoma (IMmotion151): A multicentre, open-label, phase 3, randomized controlled trial. Lancet 2019 Jun 15;393(10189):2404-2415.


