He first summarized the data from the PROfound trial, first showing a radiographic progression-free survival benefit for all metastatic castration-resistant prostate cancer (mCRPC) patients who had progressed on second-generation anti-androgen therapy and were found to have somatic deleterious mutations in certain homologous recombination repair genes or germline deleterious mutations in BRCA1 or BRCA2.1 A follow-up publication confirmed a statistically significant overall survival benefit in cohort A patients, those with deleterious alterations in BRCA1, BRCA2, or ATM.2 The Federal Drug Administration (FDA) label for olaparib, showing which genetic alterations and sample types allow its use, is shown below. An alteration was deemed deleterious if it was predicted to cause protein truncation (nonsense, consensus splice site, or frameshift alteration), was a well-described missense variant, or the presence of homozygous deletion or genomic truncating arrangement was detected. Importantly, no patients with RAD51C and FANCL deleterious alterations were enrolled in this trial. 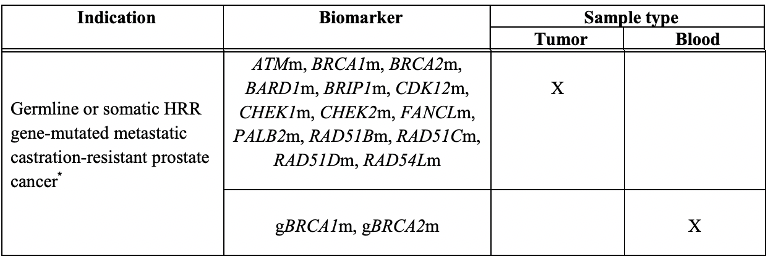
The TRITON2 trial3 demonstrated a 43.5% response with the PARP inhibitor rucaparib in mCRPC patients who had progressed on one or two second-generation anti-androgen therapies and were found to have a deleterious germline or somatic BRCA1 or BRCA2 alteration. Patients in this trial more commonly had BRCA2 alteration (102/115 patients). Deleterious alterations were identified if they were frameshift mutations, certain missense mutations, nonsense mutations, genomic rearrangements, splice site mutations, or homozygous loss that were predicted to cause protein dysfunction.
Dr. Nelson then summarized whether these approvals represent a “Sea Change”. Supporting the yes perspective, he discussed that there is evidence of a substantial clinical benefit in an advanced patient population that has limited therapies, is biomarker-driven, has changed practice, and importantly has set the foundation for refinement and improvement in the application of these therapies. Supporting the no perspective, Dr. Nelson mentioned that patients are not cured by these therapies and often have short responses, that one might have predicted this efficacy based on known efficacy in ovarian and breast cancers, that many patients who were biomarker positive did not respond to therapy, and that the indications for use may not be as precise as they should be.
Going into more detail, Dr. Nelson first described the underlying mechanism of synthetic lethality thought to underlie the efficacy of PARP inhibition in the context of mutations in homologous recombination repair. He showed the below image with citation, showing how normal cells have multiple pathways to maintain DNA sequence fidelity. In cells with deficient homologous recombination (HR), they rely on the base excision and NHEJ repair pathways, which utilize PARP proteins to repair DNA. Thus, inhibiting PARP function works in synthesis with the HR deficiency to inhibit the ability for a cell to effectively repair DNA, resulting in cell death.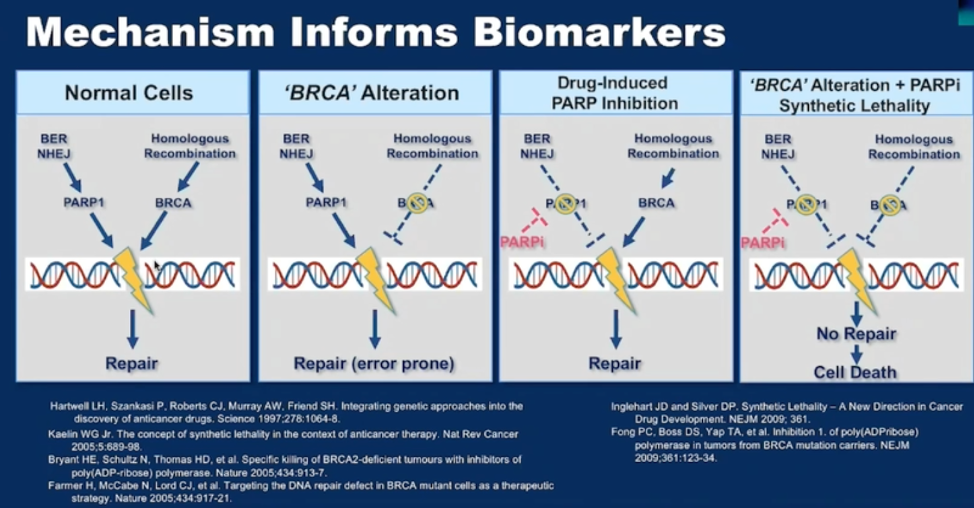
Importantly, DNA damage repair is a very complex process, and it is critical to note that alterations in the various component of HR are not equivalent in terms of their functional impact on DNA damage repair. Therefore, using specific mutations only may impact the precision and accuracy of patient selection for these drugs. Specifically, the FDA approved biomarkers for these drugs only indirectly reflect deficiencies in homologous recombination repair, are not equivalent with respect to the mechanism, there is no requirement for biallelic loss (which is thought to be the most functionally relevant), and some gene alterations on the label for olaparib do not seem to respond to therapy. By limiting the rucaparib indication to alterations in BRCA1/2, Dr. Nelson argued that this approval lacks sensitivity for identifying patients who may benefit, and by having many genes on the olaparib label, he argued that this approval lacks specificity. Using biomarkers with limited specificity and sensitivity can result in the administration of ineffective therapy, additional side effects, and significant cost.
Dr. Nelson then discussed alternative biomarkers of HR deficiency that may have better sensitivity and specificity and could result in better patient selection. These include the identification of the COSMIC mutational signature 3, identification of genomic scars associated with HR deficiency (loss of heterozygosity, large scale transitions, and telomere allelic imbalance), and other functional studies like RAD51 foci generation and other ex vivo assays for DNA repair.
Finally, he offered a case example of a patient who had circulating tumor DNA analyzed for mutations and was found to have a mutation in the ATM gene and highlighted three concerns with using this test to justify olaparib therapy. (1) ATM mutation alone does not have very strong predictive value for PARP inhibitor response, (2) There is no information provided routinely about the presence of biallelic alteration, and (3) Many alterations in HR genes in circulating DNA can be associated with clonal hematopoiesis of indeterminate potential, confounding their interpretation. Leukocyte controls should be used if using circulating tumor DNA to detect HR deficiency.
To summarize, Dr. Nelson ended by stating that PARP inhibitors are clearly a breakthrough with substantial clinical benefit in appropriately selected patients. Refinement of the assays, in part through post-approval studies, used to identify HR deficiency will be critical to best optimize the use of these new therapies.
Presented by: Peter Nelson, MD, Professor of Human Biology, Clinical Research, and Public Health Divisions, Endowed Chair for Prostate Cancer Research, Fred Hutch, University of Washington School of Medicine
Written by: Alok Tewari, MD, Ph.D., Medical Oncologist at the Dana-Farber Cancer Institute, at the 2021 American Society of Clinical Oncology Genitourinary Cancers Symposium (#GU21), February 11th-February 13th, 2021
References:
1. de Bono J, Mateo J, Fizazi K, et al., Olaparib for Metastatic Castration-Resistant Prostate Cancer. N Engl J Med; 382, (2020):2091-2102
2. Hussain M, Mateo J, Fizazi K, et al. Survival with Olaparib in Metastatic Castration-Resistant Prostate Cancer N Engl J Med; 383, (2020):2345-2357
3. Abida W, Akash P, Campbell D, et al. Rucaparib in Men With Metastatic Castration-Resistant Prostate Cancer Harboring a BRCA1 or BRCA2 Gene Alteration. J Clin Oncol. 38(32), (2020):3763-3772
Related Content:
Patients with Metastatic Castration-Resistant Prostate Cancer (mCRPC) Can Be Preselected to Maximize Benefit of Olaparib - Maha Hussain


