(UroToday.com) On the second day of the American Society for Clinical Oncology (ASCO) Genitourinary Cancer Symposium 2023 focussing on urothelial cancer, the Rapid Abstract Session included a presentation from Dr. Kyle Rose discussing results of a prospective study assessing the role of cell-free urinary tumor DNA (cfDNA) prior to repeat transurethral resection of bladder tumor (TURBT) for patients with non–muscle-invasive bladder cancer (NMIBC).
There is an unmet need to understand the need for re-resection for patients initially diagnosed with NMIBC. One potential approach is cell-free urinary tumor DNA (utDNA) generated from tumor genomic sequencing. This emerging biomarker has demonstrated potential to detect minimal residual disease (MRD) in muscle-invasive and metastatic bladder cancer. In this study, the authors performed a prospective study to assess utDNA’s ability to measure MRD at the time of standard-of-care repeat transurethral resection of bladder tumor (rTURBT) in non-muscle invasive bladder cancer (NMIBC).
To do so, the authors enrolled patients with high-risk NMIBC were enrolled prior to rTURBT. Both the index tumor and rTURBT mutational profile underwent whole exome sequencing using PredicineWES across 20,000 genes. They reported genes with non-synonymous (NS) mutations observed in index and rTURBT specimens identified in a minimum of two samples.
Urine samples were taken immediately prior to r-TURBT. utDNA detection was performed by ultra-deep sequencing of urinary cfDNA using a custom panel of up to 50-baseline mutations per patient together with a fixed core panel covering hotspot regions and actionable variants via PredicineBEACON. Urinary tumor fraction was used to call utDNA positivity. The primary endpoint was the detection of utDNA to predict MRD at the time of repeat-TURBT.
The authors enrolled 11 patients who underwent rTURBT for high-risk NMIBC. Residual tumor was detected at rTURBT in eight of 11 (73%) patients. Fifty genes with NS mutations found in at least two samples were identified including TP53, PIK3CA, RB1, MYC, CDKN1A and ARID1A. A median of 146 (range 39-418) and 91 NS (range 2-312) mutations were identified in the index and re-TUR specimens, respectively.
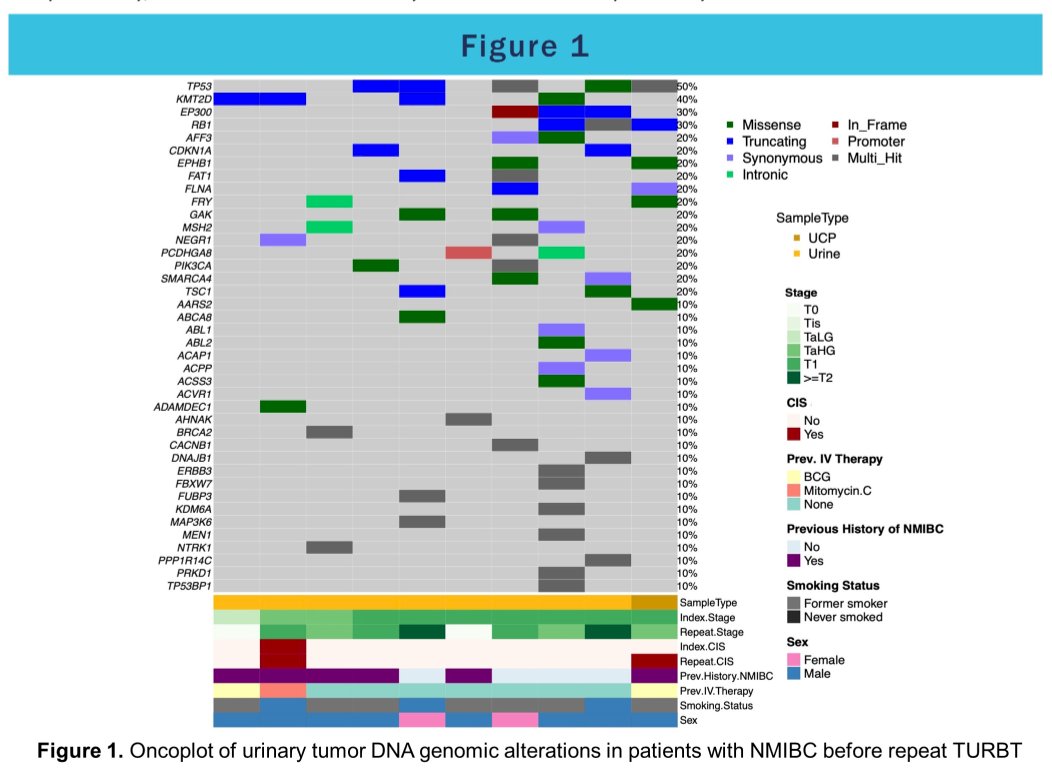
The authors found concordance rates of 83% on average between index and rTURBT, highest for patients upstaged to T2 disease, and lowest for patients harboring CIS. New somatic variants not observed in the index tissue were detected in rTURBT for 4/6 patients and 7/14 were also detected in the pre-operative urine.
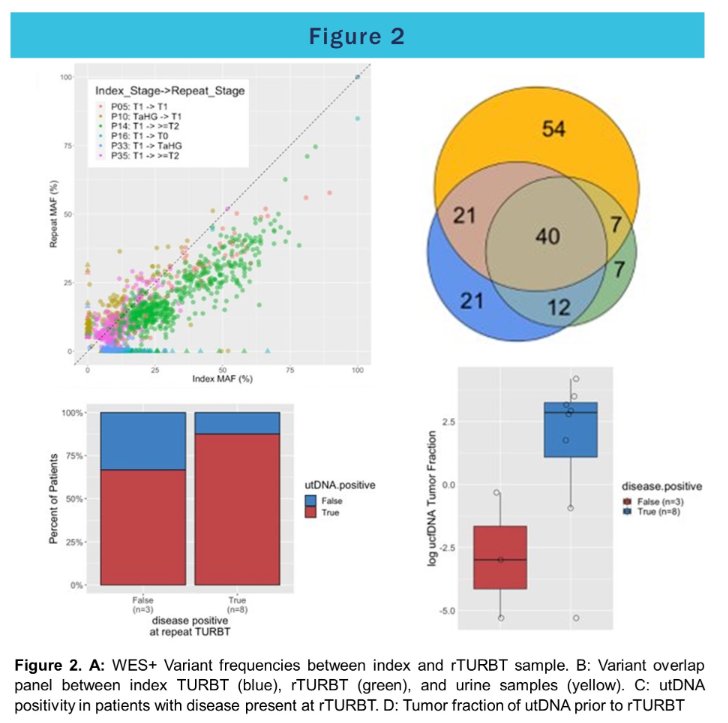
Overall, 50% of utDNA mutations were concordant with index tumor tissue, 39% were concordant with rTURBT tissue, and 33% were concordant with both. The tumor fraction of utDNA was higher in patients with residual disease (mean 2.5% vs. 0.2%, p=0.10). Using tumor-fraction to predict MRD, the area under the receiver-operator curve was 0.85. Using an optimal threshold of ≥3.3% tumor-fraction to define utDNA positivity, the test had a sensitivity of 75% at 100% specificity.
Thus, the authors concluded that urinary tumor DNA shows promise as a surrogate for minimal residual disease and may predict TURBT pathology for NMIBC. Further, they found that genomic alterations between index and rTURBT tumors were highly concordant in papillary tumors even in the setting of upstaging, which may aid in targeted intravesical or systemic therapy selection. However, larger cohorts and long-term follow up are needed to determine if utDNA can risk-stratify patients prior to rTURBT and predict long-term recurrence.
Presented by: Kyle M. Rose, MD, Moffitt Cancer Center, Tampa, FL


