(UroToday.com) The 2024 American Society of Clinical Oncology Genitourinary (ASCO GU) cancers symposium held in San Francisco, CA between January 25th and 27th was host to a prostate cancer rapid oral abstract session. Dr. Umang Swami presented the results of an analysis of differences in the genomic, transcriptomic, and immune landscapes of prostate cancer, based on the site of metastasis.
Dr. Swami noted the contemporary distribution of metastatic sites in advanced prostate cancer, with the most common sites of metastasis being the bone (84 – 91%), followed by distant lymph nodes (8.7 – 10.6%), liver (4.5 – 10.2%), and thorax (5.7 – 9.1%).1

The site of metastasis is of utmost clinical prognostic significance, with patients harboring visceral metastases, particularly liver metastases, having a significantly worse overall survival outcomes, compared to those with other sites of disease spread.2 However, to date, differences in genomic, transcriptomic, and immune profiles of prostate cancer lesions by metastatic site that drive these clinical outcomes differences remain unknown.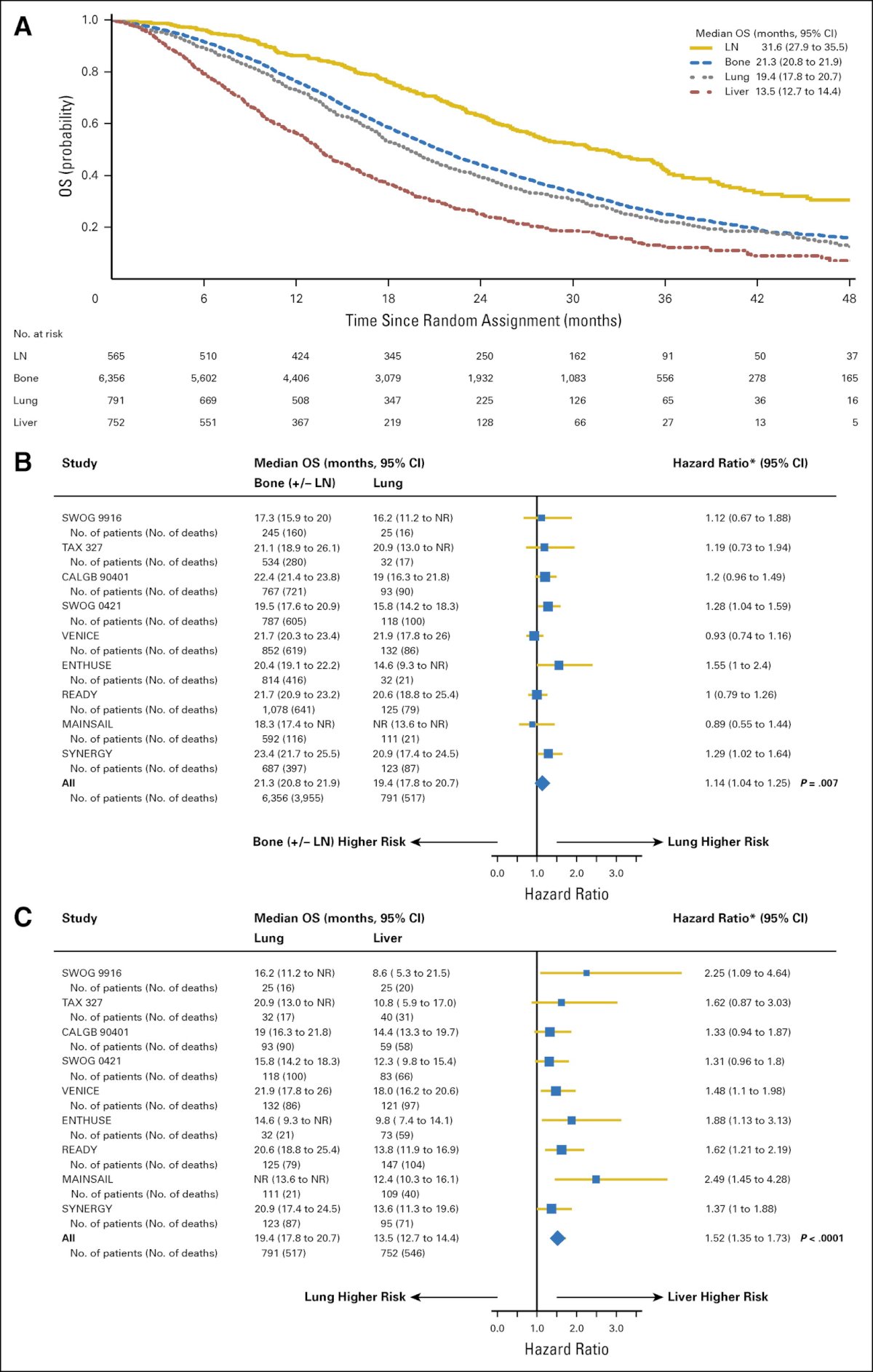
The primary objective of this study was to comprehensively characterize the differences in molecular and immune landscape of prostate cancers, based on metastatic site utilizing a large, multi-institutional, real-world dataset of patients with prostate cancer (Caris Life Sciences, Phoenix, AZ).
The authors performed DNA (592-gene panel or whole exome) and RNA (whole transcriptome) sequencing for the primary prostate cancer and unpaired metastatic sites. Immunohistochemistry and next-generation sequencing were used to test for mismatch repair deficiency (dMMR) and microsatellite instability-high (MSI-H) statuses. The investigators also assessed the tumor mutational burden (TMB high defined as >10 mut/Mb). Androgen receptor (AR) signaling score and Neuroendocrine Prostate Cancer (NEPC) score were calculated using previously defined RNA-based signatures. Pathway enrichment was determined by Gene Set Enrichment Analysis (GSEA). Immune cell fractions were inferred by deconvolution of whole transcriptome sequences using quanTIseq. Statistical comparisons were performed using the Chi-square and Mann-Whitney U tests.
The analysis included specimens from 6,074 patients. The most common specimen sites were the prostate (56.2%), followed by lymph node (14%), bone (12.9%), and hepatic (7.7%) sites of metastases.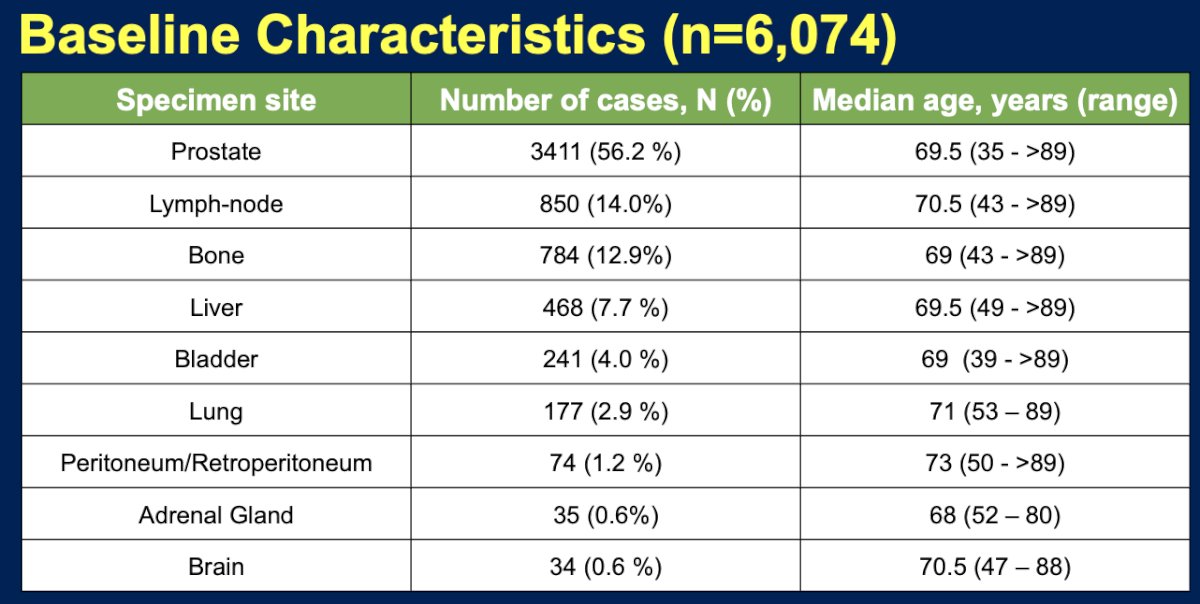
Compared to primary prostate cancer:
- Lesions from liver metastases demonstrated a significantly higher frequency of Retinoblastoma 1 (RB1), Adenomatous Polyposis Coli (APC), and AR mutations, but less frequent Speckle-type POZ protein (SPOP) mutations
- Lesions from lung metastases demonstrated significantly higher frequencies of SPOP, Kristen rat sarcoma (KRAS), PIK3CA, APC, and BRCA-1 associated protein 1 (BAP1) mutations
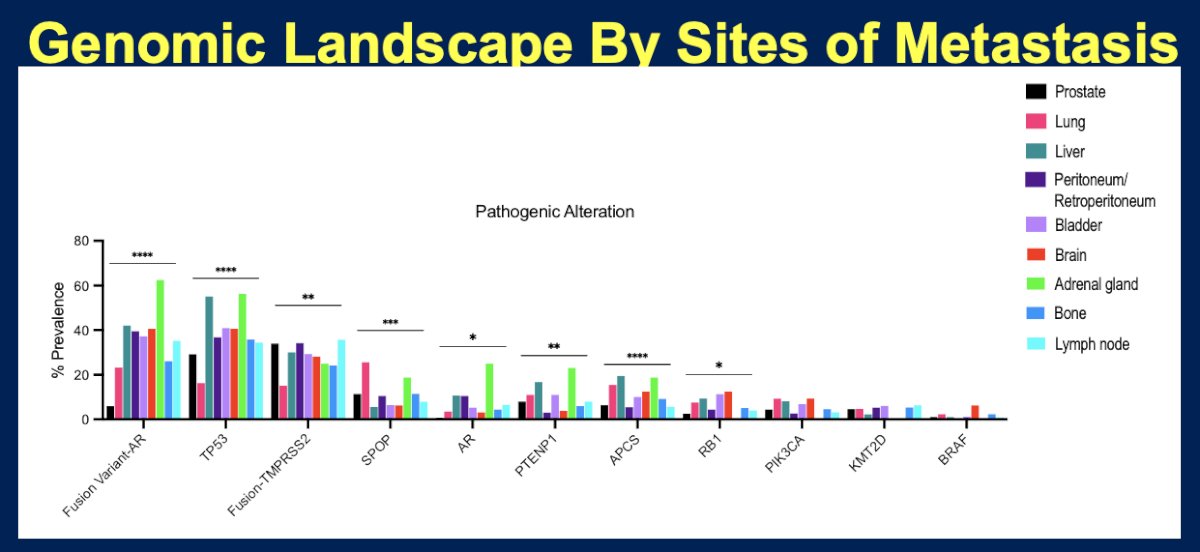
Compared to primary prostate cancer, metastatic sites had higher frequencies of AR-V7 variant mutations.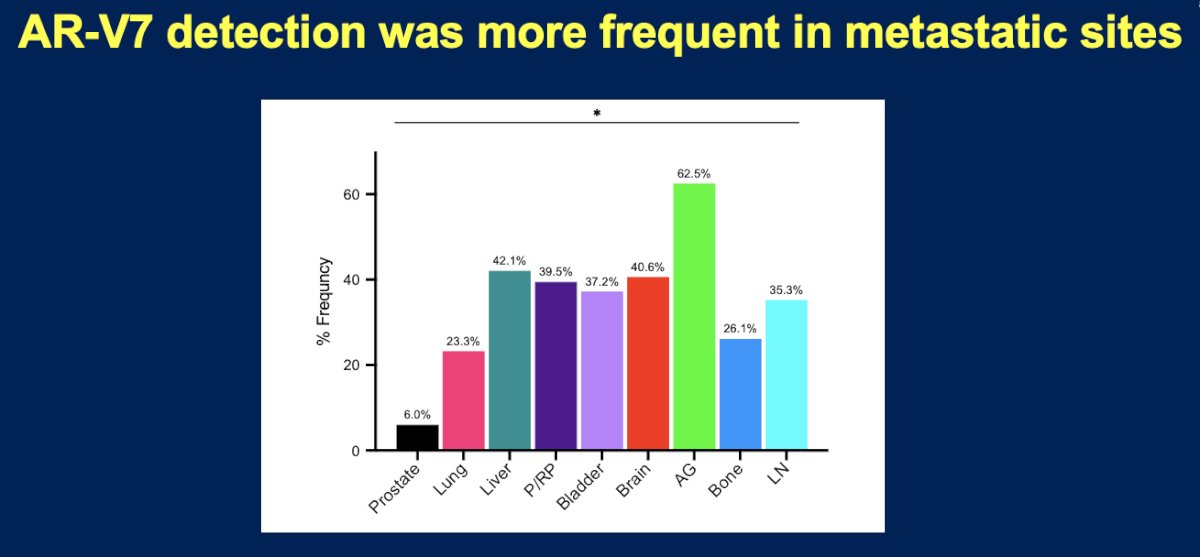
Metastatic sites were similar to primary prostate cancer lesions with regards to TMB-high and dMMR/MSI-H alteration frequencies.
With regards to the tumor microenvironment, compared to primary prostatic lesions, the tumor microenvironment of liver metastatic lesions was less enriched with macrophages M2, NK cells, Tregs, B cells, and neutrophils (p-value<0.05). The tumor microenvironment of lung lesions was similarly less enriched with NK cells, but more highly enriched with Tregs (p-value <0.05).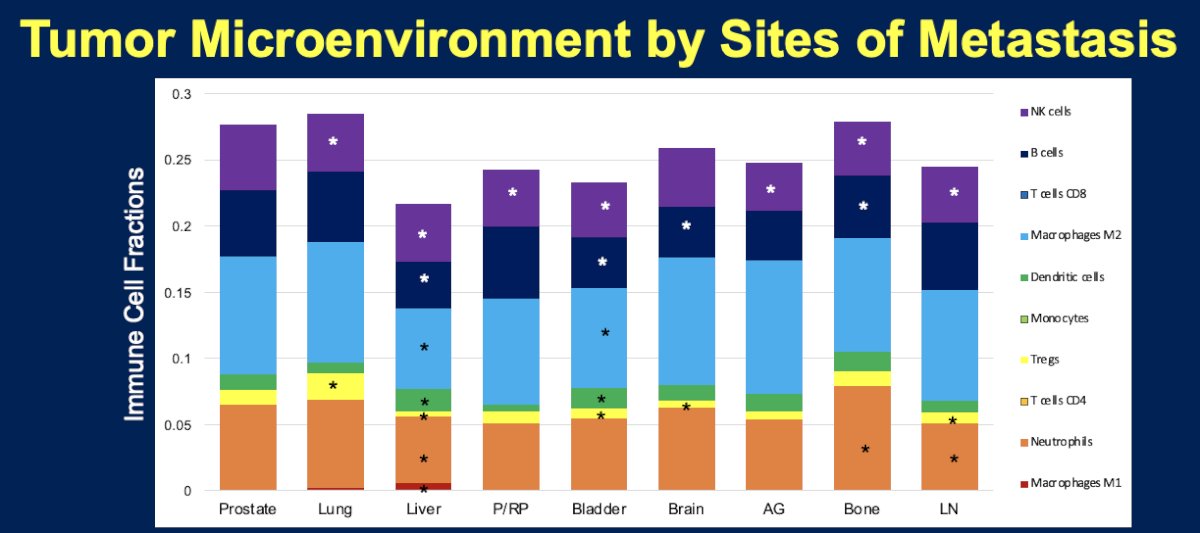
With respect to RNA signature scores, compared to primary prostatic lesions:
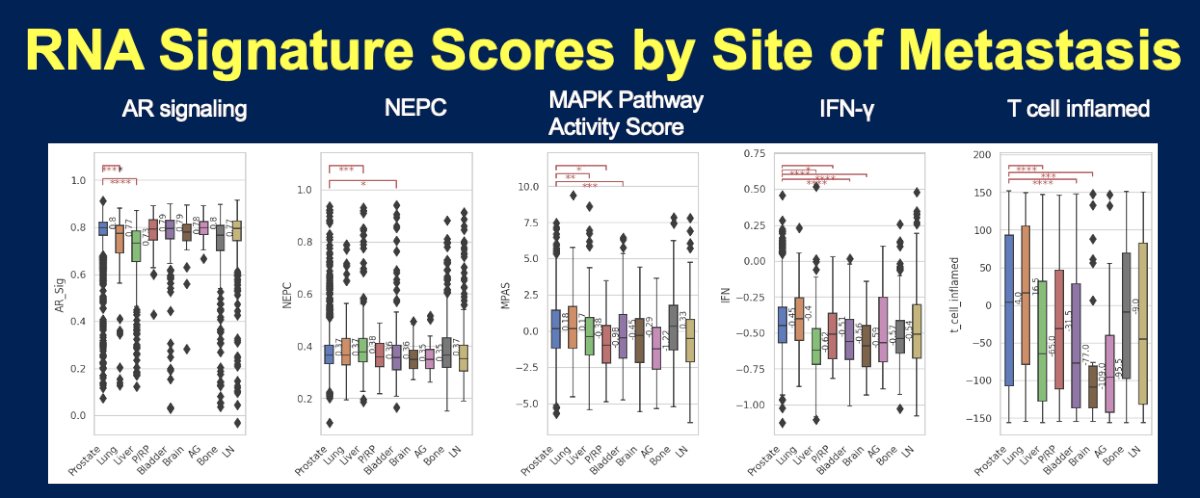
- Liver metastatic lesions demonstrated lower AR signaling, MAPK Pathway Activity score, T-cell inflamed, and IFN-γ signatures, but higher NEPC scores
- Lung metastatic lesions demonstrated lower AR signaling scores
Pathway enrichment analysis using GSEA demonstrated that E2F, G2M checkpoint, and MYC target pathways were significantly more enriched in both visceral and non-visceral metastasis (p<0.05) compared to primary prostate cancer lesions.
Dr. Swami concluded that compared to primary prostate cancer lesions:
- Liver metastatic lesions demonstrate a significantly higher frequency of RB1, APC, and AR mutations, less frequent SPOP mutations, lower AR signaling, T-cell inflamed and IFN-γ signatures, and higher NEPC scores. The tumor microenvironment was less enriched with macrophages M2, NK cells, Tregs, B cells, and neutrophils (p<0.05).
- Lung metastatic lesions demonstrate a significantly higher frequency of SPOP, KRAS, PIK3CA, APC, and BAP1 mutations. The tumor microenvironment was less enriched with NK cells, but more highly enriched with Tregs.
- Metastatic sites were similar in TMB-high and dMMR/MSI-H frequencies
- E2F, G2M checkpoint, and MYC target pathways are more upregulated in both visceral and non-visceral metastasis, compared to primary prostate cancer lesions.
- These findings elucidate molecular, immunologic, and tumor microenvironment differences between primary and metastatic sites in patients with advanced prostate cancer.
- Understanding the biology of metastatic tropism to specific sites will help explain the reasons for differential outcomes based on sites of metastasis and may facilitate future drug development.
Presented by: Umang Swami, MD, MS, Assistant Professor, Division of Oncology, Department of Internal Medicine, Huntsman Cancer Institute at the University of Utah, Salt Lake City, UT
Written by: Rashid Sayyid, MD, MSc – Society of Urologic Oncology (SUO) Clinical Fellow at The University of Toronto, @rksayyid on Twitter during the 2024 American Society of Clinical Oncology Genitourinary (ASCO GU) Cancers Symposium, San Francisco, CA, January 25th – January 27th, 2024
References:- Gandaglia G, Karakiewicz PI, Briganti A, et al. Impact of the Site of Metastases on Survival in Patients with Metastatic Prostate Cancer. Eur Urol. 2015;68(2):325-34.
- Halabi S, Kelly WK, Ma H, et al. Meta-Analysis Evaluating the Impact of Site of Metastasis on Overall Survival in Men With Castration-Resistant Prostate Cancer. J Clin Oncol. 2016;34(14):1652-9.


