(UroToday.com) Despite advances in biomarker development, early detection of aggressive prostate cancer (Gleason Group 3-5) continues to pose significant clinical challenges. Several authors involved in this study previously developed the Michigan Prostate Score for individualized risk prediction of aggressive prostate cancer.1 The Michigan Prostate Score uses transcription-mediated amplification to quantify expression of TMPRSS2:ERG and PCA3 from whole urine obtained after a digital rectal exam, combined with serum PSA. To improve upon the Michigan Prostate Score, Dr. Salami and colleagues at the AUA 2020 virtual annual meeting presented results of their study describing the pre-clinical development and validation of a targeted next-generation RNA sequencing assay Michigan Prostate Score.
For this study, the authors selected patients with available Michigan Prostate Score scores, as well as representing a spectrum of disease grade on biopsy (benign to Grade Group 5). 2.5 mL of post-digital rectal exam whole urine was used to assess approximately 90 prostate transcriptomic biomarkers, including the transmembrane protease serine 2:v-ets erythroblastosis virus E26 oncogene homolog (TMPRSS2:ERG), Prostate cancer antigen 3 (PCA3), and additional isoforms of common prostate gene fusions, mRNAs, lncRNAs, and expressed mutations. The primary analytic cohort was an extreme case design (Gleason Group 3-5 vs benign/Gleason Group 1) and a secondary analytic cohort (active surveillance cohort).
Next-generation RNA sequencing assay Michigan Prostate Score showed a 98% informative sample rate, high technical reproducibility, robustness and concordance with orthogonal methods (TMA and RT-qPCR), and was able to detect an expressed HOXB13 p.G84E variant and SPOP mutations: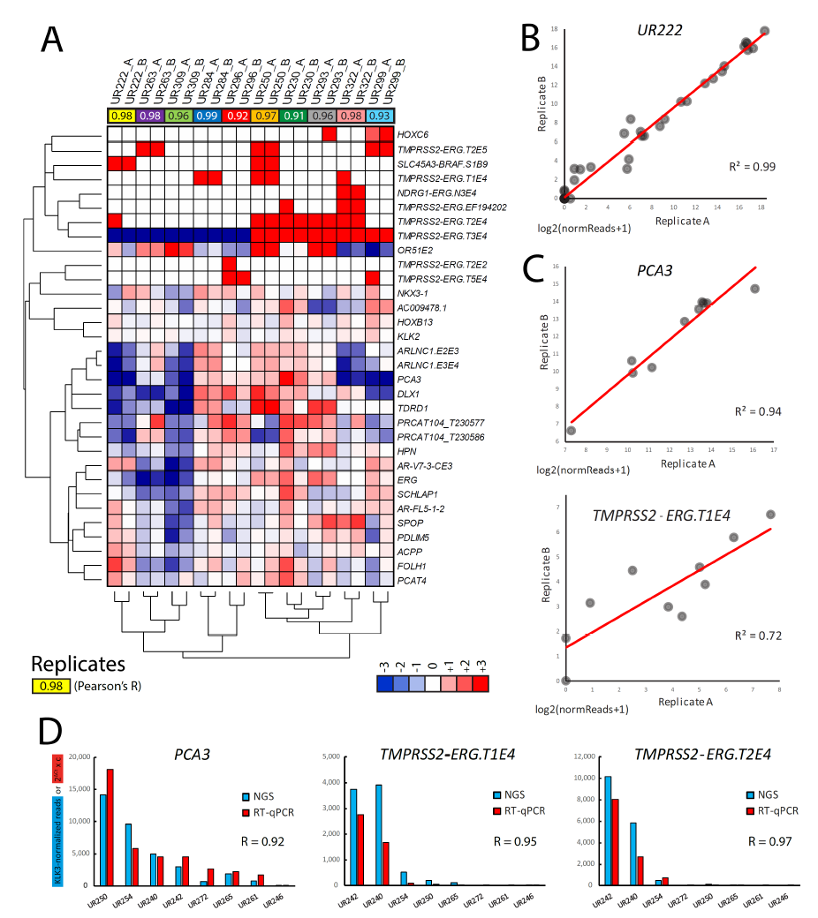
Next-generation RNA sequencing assay Michigan Prostate Score accurately recapitulated clinical Michigan Prostate Score-measured risk scores for presence of prostate cancer or high-grade prostate cancer (Gleason Score >6) on biopsy as determined by the clinical Michigan Prostate Score versus the same model but with next-generation RNA sequencing assay Michigan Prostate Score data:
In an extreme design cohort (benign or Gleason 6 versus Gleason = 4+3=7 cancer) next-generation RNA sequencing assay Michigan Prostate Score showed expected differences in the levels of TMPRSS2:ERG T1E4 (p<0.00001) and PCA3 (p=0.02), with additional TMPRSS2:ERG splice isoforms and other biomarkers also showing significantly different expression between low versus high-grade cancer. The authors used a machine learning approach trained on a subset of the extreme design cohort (n=73) to generate a 29-transcript model that outperformed the Michigan Prostate Score and serum PSA in two validation cohorts:
1. A held-out set from the extreme design cohort n=36, (AUC 0.82 versus 0.73 and 0.69, respectively)
2. A separate prostate cancer active surveillance cohort n=45, (AUC 0.66 vs. 0.58 and 0.53, respectively)
Dr. Salami and colleagues noted several limitations with the current study including (i) that this was a selected cohort to represent the extremes of prostate cancer grade groups; further prospective development and validation in clinically practical settings is warranted, and (ii) whole urine was collected after an attentive digital rectal exam, which is necessary for assay testing; exploration of pre-digital rectal examination urine is ongoing.
Dr. Salami concluded his presentation with the following take-home messages:
- The novel targeted RNA next-generation RNA sequencing assay Michigan Prostate Score assay can detect transcriptomic and expressed genomic alterations in post-digital rectal exam urine
- These results support the potential utility and continued development of the urine-based targeted next-generation sequencing assay to supplement serum PSA for improved early detection of aggressive prostate cancer
Presented by: Simpa Salami, MD, MPH, Urologic Oncologist and Assistant Professor of Urology at the University of Michigan.Urology University of Michigan, Ann Arbor, Michigan
Co-Authors: Andi K. Cani, Kevin Hu, Javed Siddiqui, Yingye Zheng, Sumin Han, Chia-Jen Liu, Daniel H. Hovelson, Srinivas Nallandhighal, Trinh Pham, Lanbo Xiao, Heng Zheng, Jeffrey J. Tosoian, Ganesh S. Palapattu, Todd M. Morgan, Aaron Udager, John T. Wei, Arul M. Chinnaiyan, Scott A. Tomlins
Written by: Zachary Klaassen, MD, MSc – Assistant Professor of Urology, Georgia Cancer Center, Augusta University/Medical College of Georgia, Twitter: @zklaassen_md at the 2020 American Urological Association (AUA) Annual Meeting, Virtual Experience #AUA20, June 27- 28, 2020
Reference:
1. Salami SS, Schmidt F, Laxman B, et al. Combining urinary detection of TMPRSS2:ERG and PCA3 with serum PSA to predict diagnosis of prostate cancer. Urol Oncol 2013;31:566-571.


