(UroToday.com) Mr. David Gelikman of the National Cancer Institute (NIH) gave a fascinating podium talk on the integration of artificial intelligence (AI) assistance in interpreting biparametric magnetic resonance imaging (bpMRI) for prostate cancer (PCa) analysis. Currently, there exists significant inter-reader variability in bpMRI interpretation among clinicians. With technological advancements, AI has emerged as a promising tool to mitigate this variability by enhancing accuracy and consistency in diagnostic imaging interpretation. Given these challenges, Mr. Gelikman and his team investigated the efficacy of a deep-learning AI model in assisting radiologists with diverse experience levels in identifying patients with PCa, interpretation of prostate bpMRI scans, index lesion size agreement, and index lesion Prostate Imaging Reporting and Data System (PI-RADS) score agreement.
The study consisted of six participating radiologists of whom three were general body readers and the other three were prostate-focused readers (completing greater than 1,500 prostate scans annually). A total of 180 total patient scans were utilized in this investigation. Of these, 120 cases consisted of whole mount pathology while the remaining 60 served as controls (consisting of benign systematic biopsy and PI-RADS 1). Using a balanced incomplete block design, patients were randomized to readers, with each reader analyzing 80 cases and 40 controls (120 in total per reader to decrease reader burden). Each MRI was read by exactly four readers. The study consisted of two phases, each phase with six blocks (20 scans per block). Once the readers were separated into two groups, phase 1 began. In this phase, each reader reads all 120 scans assigned to them. Group 1 started off with AI-assisted scans whereas Group 2 did not. Of note, only half of the scans were AI-assisted in phase 1. Once the scans were complete, a 4-week washout period took place prior to starting phase 2. In this second phase, each reader read all 120 scans again in randomized order to prevent any bias. AI-assistance was applied to scans lacking it in phase 1 and vice versa.
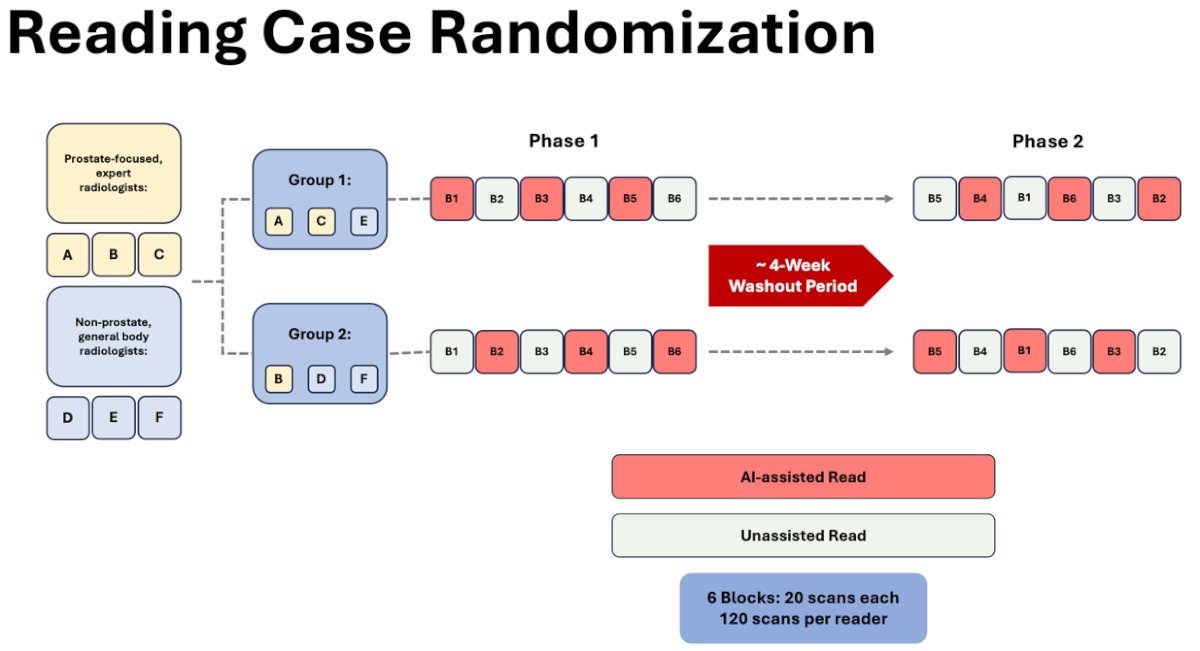
A comprehensive patient-level analysis was conducted to assess the comparative effectiveness of lesion detection at bpMRI with and without AI, examining sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and accuracy. Additionally, inter-reader agreement was evaluated on lesion measurement and PI-RADS scores.
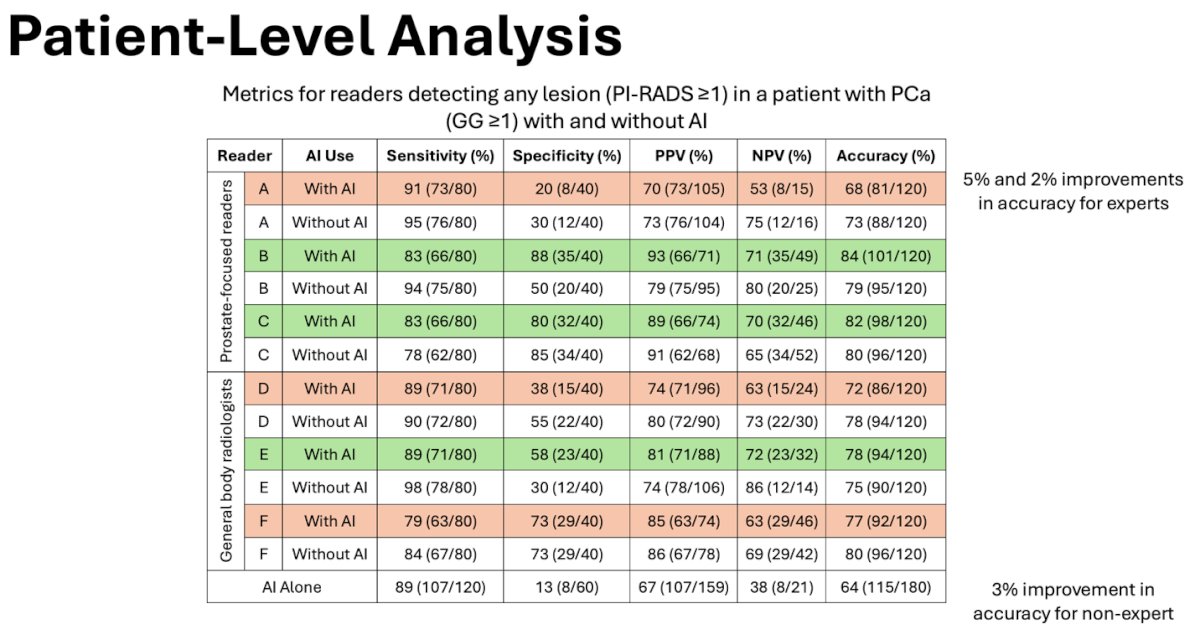
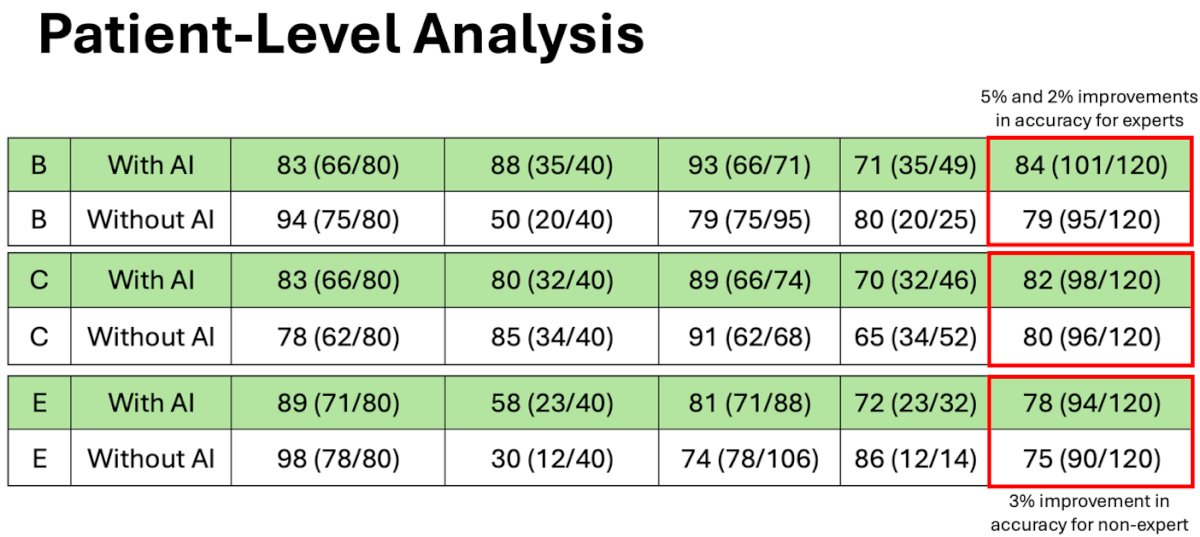
The median age of the patient cohort was 63 years (interquartile range [IQR], 57-68), with a median prostate-specific antigen level of 7.1 ng/mL (IQR, 5.1-10.8). Two of prostate-focused readers (B and C) and one of the general body radiologists (E) two experienced improvement and overall accuracy with AI-assistance of 5%, 2%, and 3%.
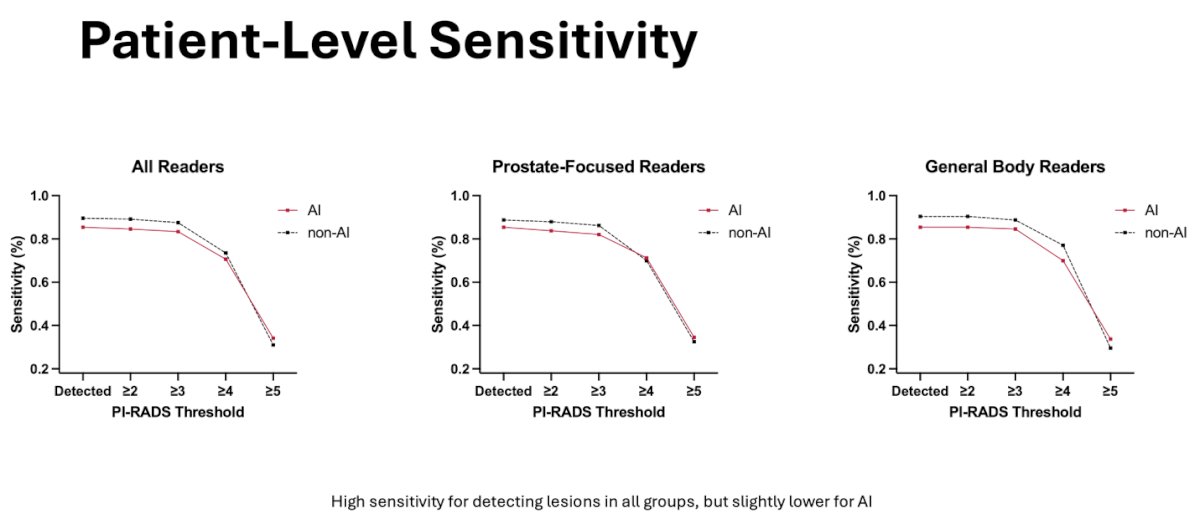
In respect to patient-level sensitivity, sensitivity for both AI and non-AI measurements was high (above 80%) for PI-RADS thresholds between ≥1-≥3 but overall decreased as the PI-RADS threshold increased. Even so, AI assessments were slightly less sensitive for all readers compared to non-AI assessments.
In respect to patient-level specificity, specificity was higher for all readers with AI assessments compared to non-AI assessments. There was a greater observed improvement among the prostate-focused readers.
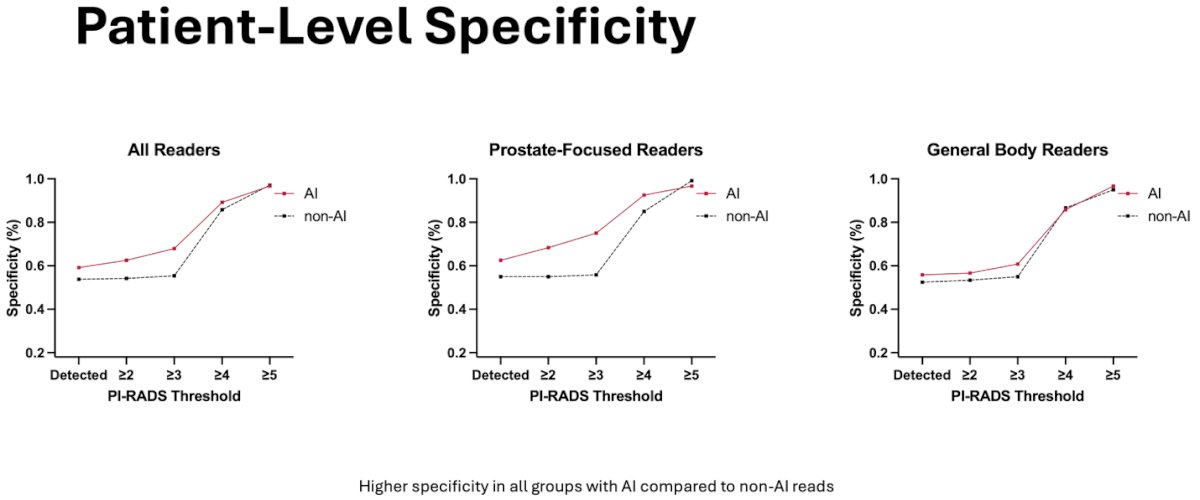
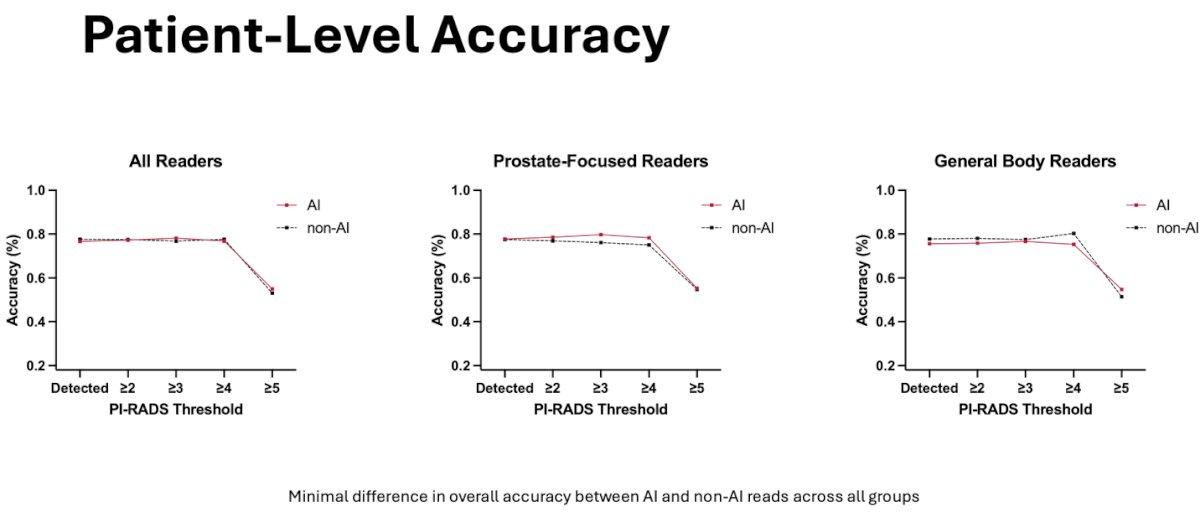
In respect to patient-level accuracy, there was no difference between AI and non-AI measurements for all readers. There was a slight observed improvement for prostate-focused readers however the difference was minimal in overall accuracy.
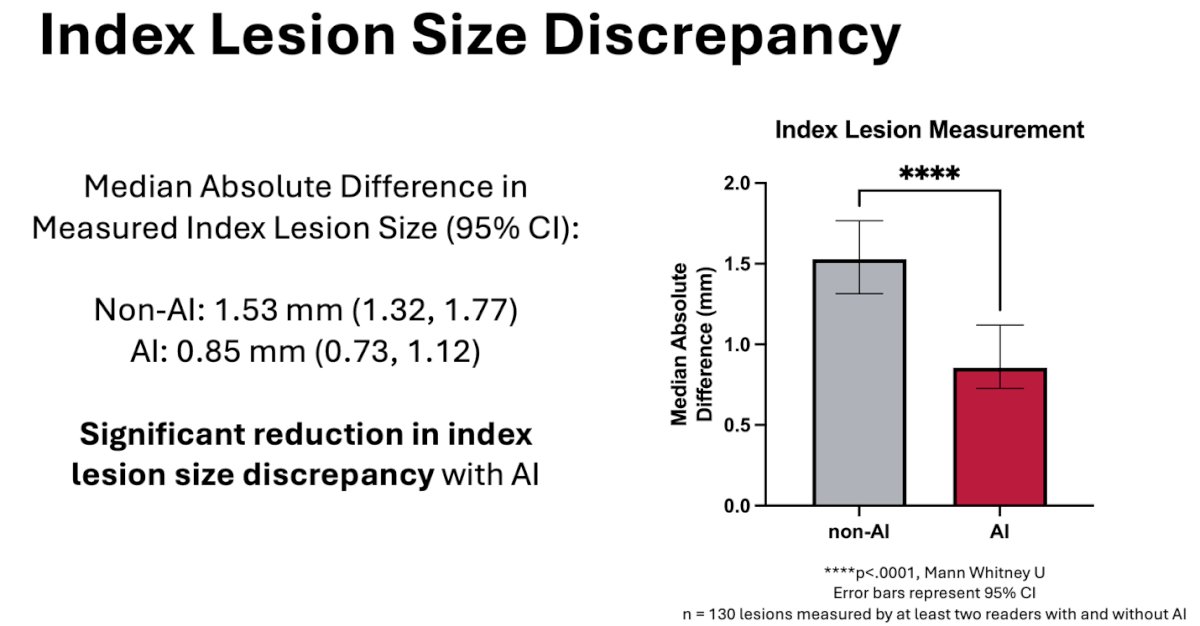
Further analysis was done for the index lesion size discrepancy among the readers to measure the difference the readers were making across the same index lesions. When using AI, the median absolute difference in the measured index lesion size was significantly reduced.
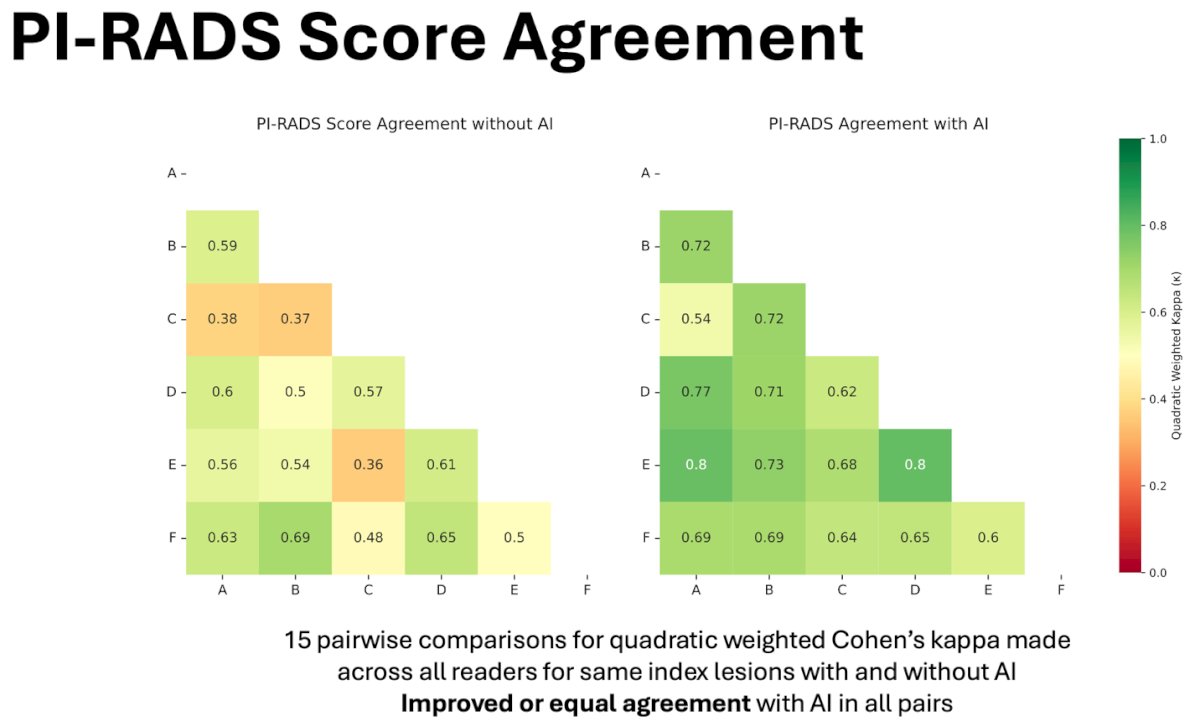
A pairwise comparison among all readers was conducted to assess agreement on PI-RADS scores, including fifteen comparisons to calculate weighted Cohen’s kappa for the same index lesions, both with and without the assistance of AI. The analysis showed an increased improved/equal agreement in all pairs with AI-assistance.
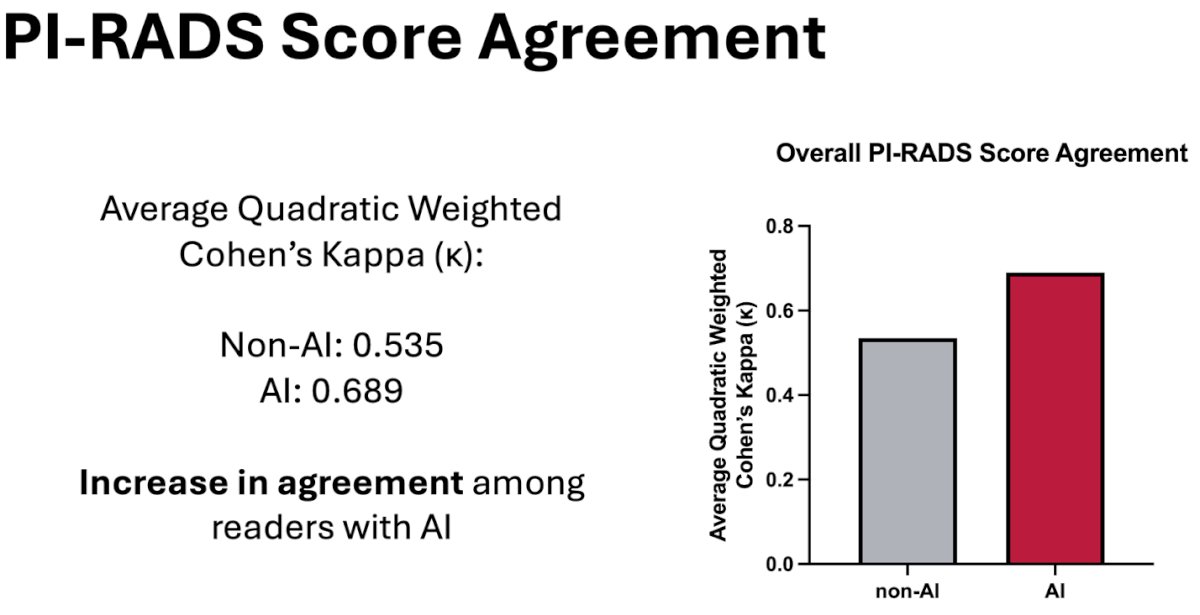
To conclude, Mr. Gelikman highlighted that his team’s AI model demonstrated the potential to enhance accuracy for select readers, although, on average, it led to decreases in sensitivity and enhancements in specificity across the majority of readers. Moreover, a notable decrease was observed in the discrepancy of measured index lesion sizes. Additionally, there was an evident improvement in the agreement of PI-RADS scores among radiologists. Mr. Gelikman ended by stating that further investigation is warranted to unveil the lesion-level sensitivity of readers with and without the aid of AI.
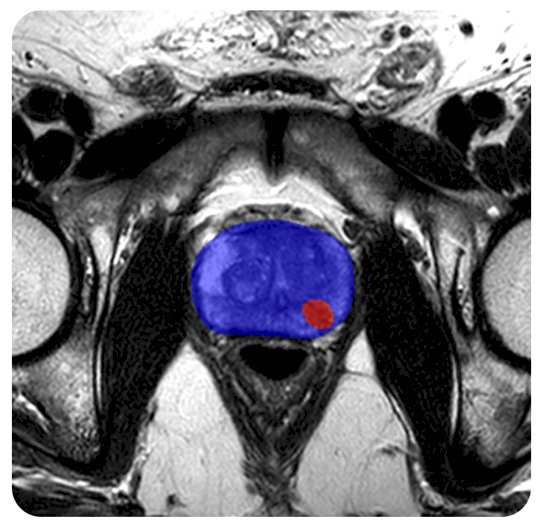
During the question-and-answer session, a member of the audience inquired about the correlation between AI reads and actual biopsy scores, particularly regarding improvements in PI-RADS scores, notably with scores 2 and 3, post-AI integration. Mr. Gelikman responded by highlighting ongoing efforts at his institution to conduct lesion-level analyses, comparing pathological results with reader analyses. Another audience member sought clarification on the AI model employed in the study and the level of assistance provided to readers during unassisted scans. Mr. Gelikman elaborated on the use of the 3D nnU-net architecture, developed and trained in-house using biparametric data from 2015-2019 on over 1,400 patients from 2015-2019. For unassisted reads, Mr. Gelikman explained that radiologists received T2, DWI, and ADC data, while AI-assisted reads included the same three factors in addition to overlay images for enhanced visualization (similar to the image below).
A subsequent query from the audience focused on the inclusion of sensitivity in the study. Mr. Gelikman clarified that while sensitivity was considered, the study didn't specifically target lesion-level sensitivity. He elaborated that the study was structured as a first-reader design rather than a concurrent reader study. With AI assistance, readers were instructed to annotate only lesions identified by the AI, thus being constrained by the sensitivity of the AI itself. A final question by the audience focused on trends observed in PI-RADS agreement analysis. Mr. Gelikman recognized the limited duration they've had the data and admitted they have not analyzed trends yet. However, he assured the audience they intend to explore trends thoroughly during the study's publication preparation.
Presented by: David Gelikman, National Cancer Institute, NIH
Written by: Seyed Amiryaghoub M. Lavasani, B.A., University of California, Irvine, @amirlavasani_ on Twitter during the 2024 American Urological Association (AUA) Annual Meeting, San Antonio, TX, Fri, May 3 - Mon, May 6, 2024.


