(UroToday.com) The 2024 Bladder Cancer Advocacy Network (BCAN) Bladder Cancer Think Tank held in San Diego, CA was host to a 2022 and 2021 Bladder Cancer Research Innovation Awardees session. Dr. John Lee discussed his lab’s research efforts for identifying both genetic determinants and vulnerabilities of histologic variants of bladder cancer.
It has been demonstrated that most cancers are genotypically and phenotypically complex, and a prime example of this is muscle invasive bladder cancer.
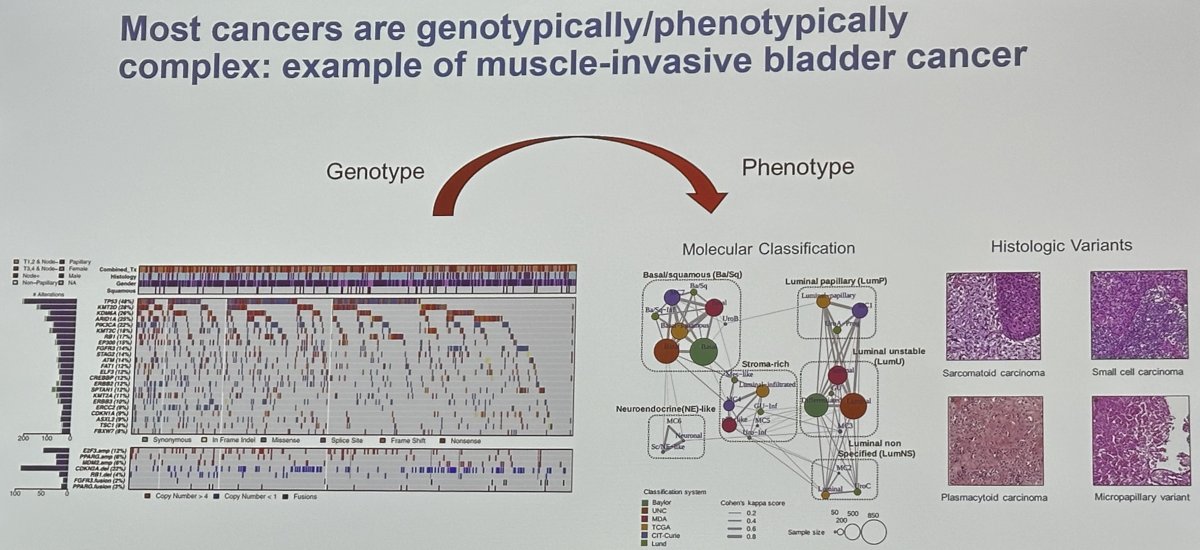
How can we more efficiently model the complexity of cancer in a genetically defined manner? Relatively few genes and gene combinations have been evaluated in genetically engineered mouse models and these certainly underrepresent the phenotypic heterogeneity of bladder cancer. Carcinogen-induced models of bladder cancer also skew heavily toward basal/squamous and are not genetically defined.
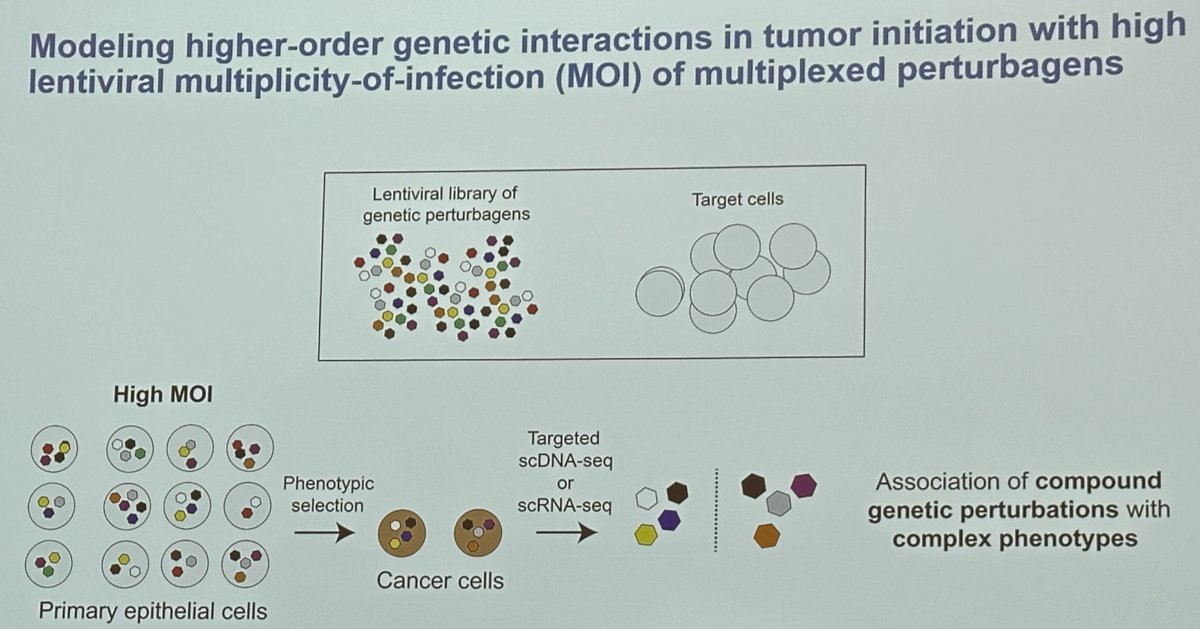
A combinatorial genetic strategy applied to an organoid transformation assay can rapidly generate diverse, clinically relevant bladder cancer models. The clonal architecture of the resultant tumors can be resolved using single-cell or spatially resolved next-generation sequencing to uncover polygenic drivers of cancer phenotypes.
Dr. Lee’s research team developed a methodology incorporating barcoded lentiviral libraries that encode cancer-associated genetic events efficiently introduced and at random into primary epithelial cells, which are engrafted in mice for tumorigenic selection, at a high multiplicity of infection (MOI). This system enables the generation of genotypically and phenotypically diverse tumors and the massively parallel single-cell lentiviral barcode sequencing of tumors to identify cooperative oncogenic drivers of malignant transformation and specific cancer phenotypes.
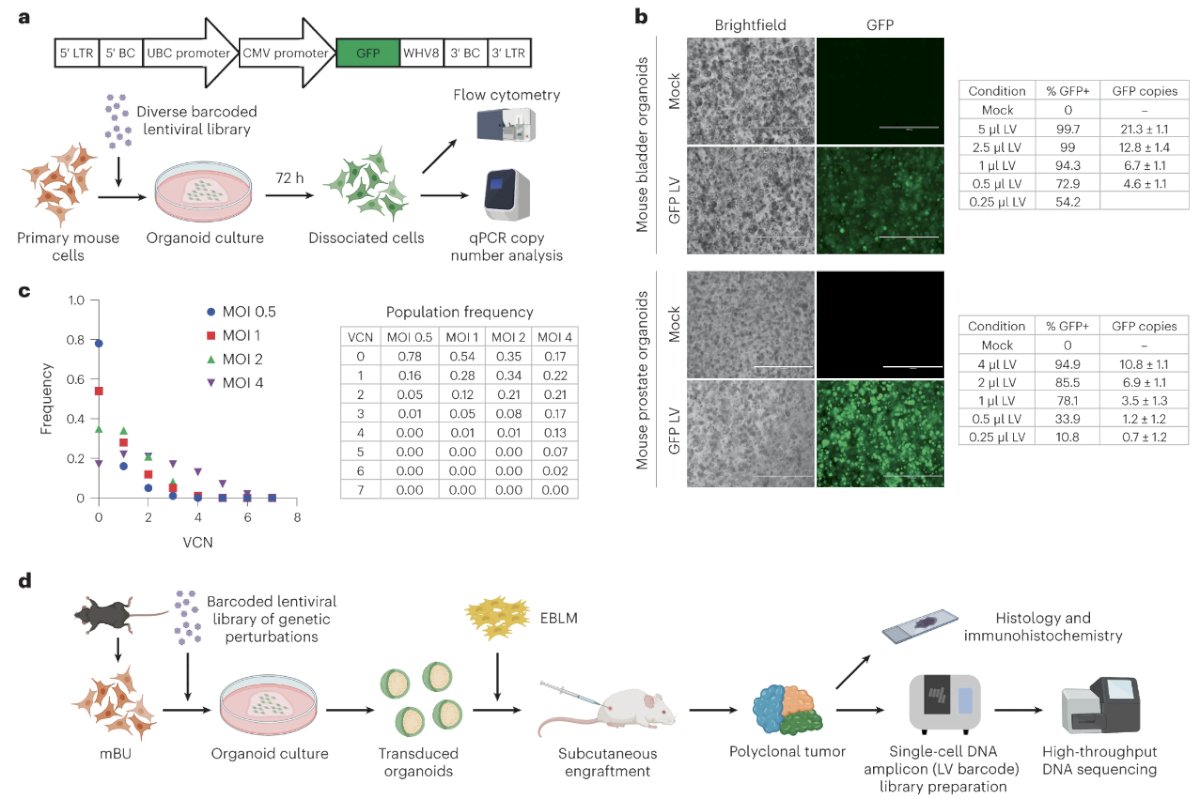
To overcome inefficient transgenesis using available lentiviral transduction protocols, Dr. Lee and colleagues theorized that enforced cell–virus contact in a constrained volume of gel matrix could increase lentiviral transduction efficiency. Primary mouse bladder urothelial and prostate epithelial cells were isolated by fluorescence-activated cell sorting. The cells were mixed into cold Matrigel containing concentrated lentivirus expressing GFP before the seeding and polymerization of organoid droplets. Near complete transduction of these cells was achieved. They next developed a barcoding system to characterize the distribution of unique proviral copies per cell. Lentiviral constructs were barcoded with matching ten-nucleotide sequences and produced as a pool. A custom single-cell amplicon panel was designed on the Mission Bio Tapestri platform to enable the sensitive enumeration of multiple uniquely barcoded lentiviruses per cell. This approach was validated using a defined population of 3T3 cells engineered with lentiviruses. Mouse prostate epithelial cells were transduced with a diverse barcoded lentiviral pool at varying MOIs, and single-cell amplicon sequencing showed relatively normal distributions of proviral copies per cell.
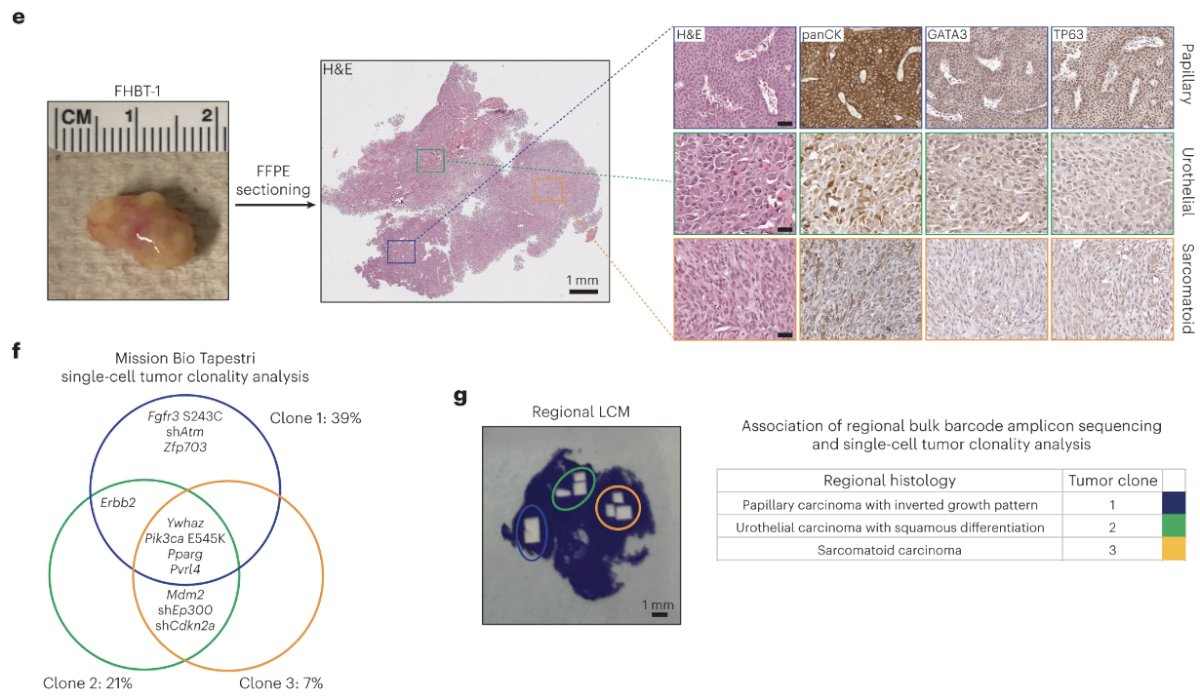
A representative tumor derived from primary mouse bladder urothelial cells transduced with the lentivirus exhibited three morphologically distinct regions consistent with papillary urothelial carcinoma with an inverted growth pattern, urothelial carcinoma with squamous differentiation and sarcomatoid urothelial carcinoma. Single-cell DNA amplicon sequencing was performed to enumerate the lentiviral barcodes to determine clonal architecture and the deconvolution of lentivirus-delivered genetic events potentially involved in tumorigenesis. Three major clones harboring distinguishable sets of lentiviral barcodes were identified; however, spatial resolution was lost secondary to single-cell dissociation. To correlate histology with clonality, they performed laser capture microdissection (LCM) of the three regions on stained tissue sections and performed bulk DNA amplicon sequencing. They found that mutant active FGFR3 signaling drives luminal papillary urothelial carcinoma.
Using this methodology, the investigators generated several tumors called the Fed Hutch Bladder Tumor (FHBT) series, which include both pure urothelial carcinoma and others with mixtures of histologic subtypes. Phenotypic characterization of the FHBT series demonstrated broad recapitulation of the cancer histologies and associated transcriptional profiles seen in the human disease.1
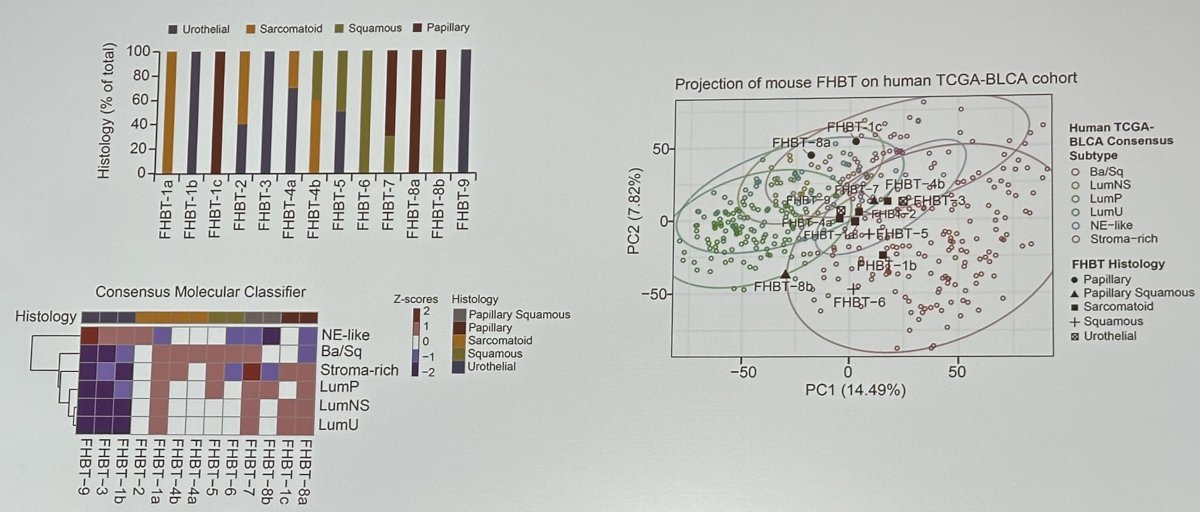
Pparg, a nuclear receptor, is downregulated in basal subtype bladder cancers that tend to be muscle invasive and amplified in luminal subtype bladder cancers that tend to be non-muscle invasive. Tate et al. have previously demonstrated that Pparg induction in urothelial tumorigenesis can result in basal progenitor cells shifting into luminal type tumors.2
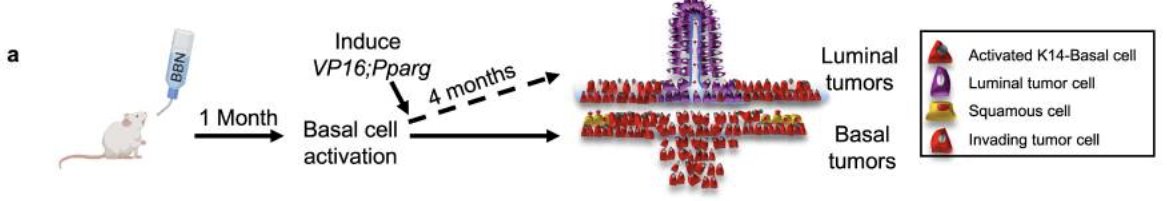
Dr. Lee’s team sought to validate Pparg constructs in mouse urothelial organoids:
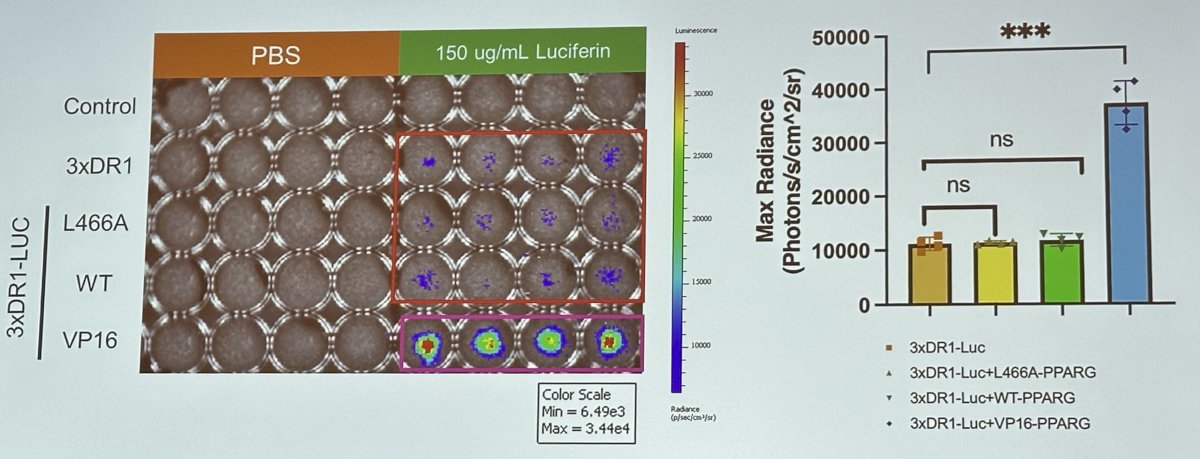
They demonstrated that the effects of Pparg variants on mouse urothelial organoid morphology and growth are consistent with the induction of luminal differentiation.
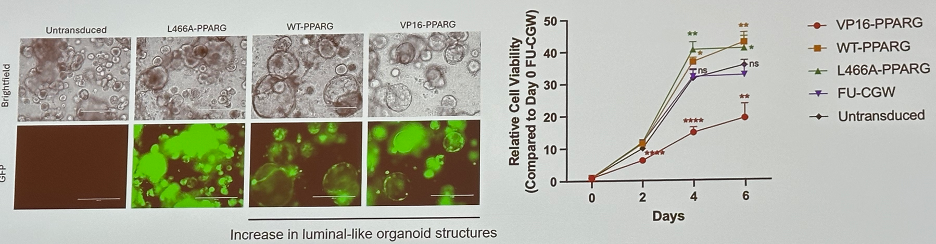
Furthermore, Pparg variants affect the efficiency of mouse urothelial organoid transformation.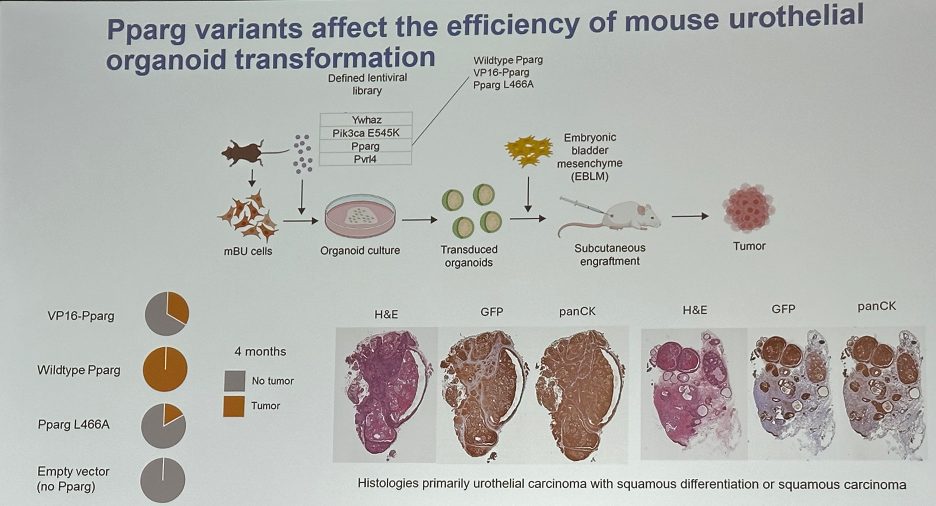
Dr. Lee summarized his presentation as follows:
- Development of a functional cancer genomics strategy to accelerate genotype-phenotype associations has been applied as proof-of-concept to bladder cancer.
- FHBT tumor models are being investigated for differential sensitivity to various therapeutic agents.
- Pparg expression appears to influence the tumorigenic potential of mouse urothelial cells.
Presented by: John K. Lee, MD, PhD, Associate Professor, Department of Medicine, Fred Hutchinson Cancer Center, University of Washington, Seattle, WA
Written by: Rashid Sayyid, MD, MSc – Robotic Urologic Oncology Fellow at The University of Southern California, @rksayyid on Twitter during the 2024 BCAN Bladder Cancer Think Tank held in San Diego, CA between August 7th and 9th, 2024
References:
- Li S, Wong A, Sun H, et al. A combinatorial genetic strategy for exploring complex genotype–phenotype associations in cancer. Nat Genet. 2024;56(3):371-6.
- Tate T, Xiang T, Wobker SE, et al. Pparg signaling controls bladder cancer subtype and immune exclusion. Nat Commun. 2021;12:6160.


