To that end, Dr. Cooperberg and colleagues, well established in the AS research sphere, describe the genomic landscape of low-risk prostate cancer to help better understand the disease, and perhaps help identify potential genomic predictors.
In this study, they work with GenomeDx, a genomic company based out of San Diego, California, which is well known for its Decipher® test. This test can be used on biopsy pathology to predict the likelihood of metastasis, high-grade disease and prostate cancer specific death for men diagnosed with localized disease. Or, using prostatectomy pathology, it can help risk stratify patients for the need for adjuvant therapy.
In this study, they analyze the biopsy pathology specimens from 473 men who were candidates for AS (stage ≤cT2N0M0, PSA≤10 ng/ml, Gleason 3+3 or low−volume 3+4), of which 408 passed quality control and were included – it should be noted that all these patients did go on to radical prostatectomy, but the RP specimens have yet to be analyzed. Affymetrix Human Exon microarray was used to generate RNA expression data. This data was compared against the data of 2043 prostatectomy specimens previously analyzed on the same platform (separate GRID cohort). Scores for 21 published prognostic signatures were calculated and gene−set enrichment analysis pathway genes were summarized to provide levels of patient risk and pathway activity.
Based on the quartiles of average scores for the 21 prognostic signature risk models, 44%, 31%, 18%, and 7%, respectively, were classified into the 1st, 2nd, 3rd, or 4th score quartiles. Importantly, clinical features were not enough to classify patients based on genomic risk. Patients in the 4th quartile: 36% were Gleason 6 disease and 42% were cT1. On the other hand, 14% of the Q1 patients were Gleason 3+4=7.

Figure below shows a heat map comparing the AS patients to the RP cohort. The top line above demonstrates that
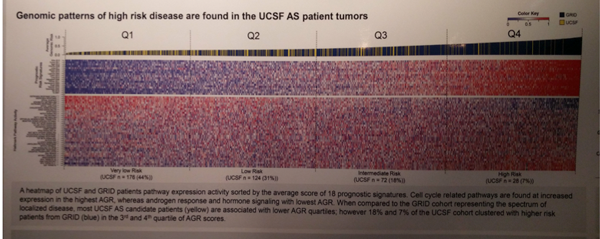
In terms of enrichment analysis, genomic risk was positively associated with cell cycle related pathway activity (E2F, G2M, MYC, DNA Repair, mTOR, mitotic spindle, p<0.001) and negatively associated with apical junction (p<0.001), epithelial−mesenchymal transition (p<0.001), and androgen receptor signaling (p<0.05) pathways.
Clustering of patients based on the expression of 36 pathways revealed two biologic groups corresponding to putative basal and luminal subtypes. Compared to higher risk RP patients, the low risk prostate cancer tumors at diagnosis were enriched for basal-like tumors (20% vs 33%, p<0.001). Of note, the luminal type had two subtypes – Luminal A and Luminal B. Luminal B has higher AGR (adjusted risk) scores, implying higher risk disease. As seen above, genomic profiles don’t always correlate with clinical features.
In summary, approximately 7% of the AS cohort had a high-risk genomic profile, indicating that there may be a subset of patients that can be identified at the outset as poor candidates for AS. As their pathway enrichment overlaps with those of high-risk prostate cancer, they may behave more like high-risk PCa. However, until clinical correlation is available, this remains just a theory. Additional assessments are required.
Limitations / Discussion Points:
Until clinical correlation is completed, this remains an exciting finding but only preliminary. Dr. Cooperberg mentioned that they are looking to eventually analyze the RP specimens
Presented by: Matthew Cooperberg
Co-authors: Erho N., Chan J., Feng F., Cowan J., Simko J., Ong K., Alshalalfa M., Kolisnik T., Margrave J., Aranes M., Du Plessis M., Buerki C., Zhao S., Tenggara I., Davicioni E., Carroll P.
Institution(s):
1. University of California, Dept. of Urology, San Francisco, United States of America
2. GenomeDx, San Diego, United States of America
3. UCSF, Dept. of Urology, San Francisco, United States of America
4. GenomeDx, San Diego, United States of America
Written by: Thenappan Chandrasekar , Clinical Fellow, University of Toronto
Twitter: @tchandra_uromd
at the #EAU17 -March 24-28, 2017- London, England


