Dr. Auffray highlighted their analysis using clinical and functional genomics data to all for the derivation of individualized tumor fingerprints and handprints. This process requires the development of a biobank from a patient cohort with the development of multi-level “-omics” data that can subsequently be integrated.

In undertaking this work, the authors developed a comprehensive approach integrating the development of raw data, its molecular analysis and integration, and clinical applications in the development of novel therapeutic agents and changes in medical practice.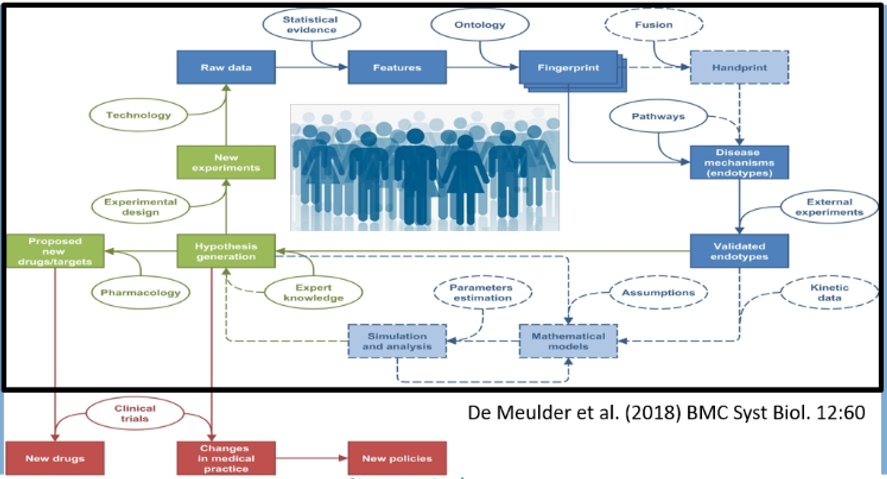
This process has led to the development of computerized disease maps for many chronic and communicable diseases which allow for an assessment of cellular interactions and biochemical mechanisms. This process involves the development of a community of experts who gather relevant data. Data are represented using systems biology graphical notation which is comprehensible to both humans and machine learning approaches.
Using wearable sensors, the group can access data on many physiologic parameters as well as common activities of daily living, constant longitudinal measurements that complement the cross-sectional nature of “-omic” data.
Dr. Auffray suggests an ongoing assessment of wellbeing for each individual using their own health as a baseline comparison using survey data, physiologic monitoring, biomarkers, DNA sequencing, serum tests, and microbiologic assays.
Such an integrative approach allows for the potential detection of disease processes prior to clinical manifestation. Such an approach is currently being leveraged in prostate cancer in the PIONEER project, coordinated by Professor James N’Dow.
Presented by: Charles Auffray, PhD, Centre National de la Recherce Scientifique, Lyon, France
Written by: Christopher J.D. Wallis, MD, PhD, Urologic Oncology Fellow, Vanderbilt University Medical Center, Nashville, Tennessee, Twitter: @WallisCJD at the Virtual 2020 EAU Annual Meeting #EAU20, July 17-19, 2020


