The 2023 European Society of Medical Oncology (ESMO) Annual Congress held in Madrid, Spain between October 20th and 24th, 2023 was host to a new diagnostic tools abstracts poster session. Dr. Jianqing Wang presented the results of a study evaluating a novel machine learning-based method for the detection of homozygous deletions of homologous recombination repair (HRR) genes in prostate cancer.
Homologous recombination comprises a series of interrelated pathways that function in the repair of DNA double-stranded breaks and inter-strand crosslinks. Accordingly, germline (i.e., inherited) HRR gene alterations, including BRCA1/2 and ATM mutations, predispose patients to an increased risk of future prostate cancer development and adverse outcomes when mutated in the germline or somatic settings. Since 2020, numerous poly ADP ribose polymerase (PARP) inhibitors have been US Food and Drug Administration (FDA) approved for the treatment of patients with germline or somatic HRR mutations:
- Rucaparib in May 2020 for mCRPC patients with deleterious Breast Cancer (BRCA) mutations who had been previously treated with an ARPI and taxane-based chemotherapy1 (TRITON2)2
- Olaparib in May 2020 for patients with deleterious or suspected homologous recombination repair (HRR) gene-mutated mCRPC with progression following prior abiraterone or enzalutamide3 (PROfound4)
- Olaparib in combination with abiraterone and prednisone (or prednisolone) in May 2023 for mCRPC patients with deleterious or suspected deleterious BRCA mutations5 (PROpel6)
- Talazoparib in combination with enzalutamide in June 2023 for mCRPC patients with HRR gene mutations7 (TALAPRO-2).8
As such, testing for and the detection of HRR deletions is critical for the provision of a personalized treatment approach for patients with advanced disease. Homozygous HRR deletions account for approximately 15% of HRR gene alterations in prostate cancer and are an important pathogenic alteration. However, the reliable detection of these homozygous deletions remains technically challenging. As such, novel methods to better identify such mutations are needed. In this study, Dr. Wang and colleagues introduced a novel machine learning-based method for the detection of homozygous deletions in formalin-fixed paraffin-embedded prostate cancer tissues.
To this end, the authors conducted approximately 30,000 homozygous and non-homozygous simulation events of varying tumor cellularity and fragment size using the sequencing data of matched tumor and wild type cell lines. These were tested using the AmoyDx HRD Complete Next Generation Sequencing (NGS) panel.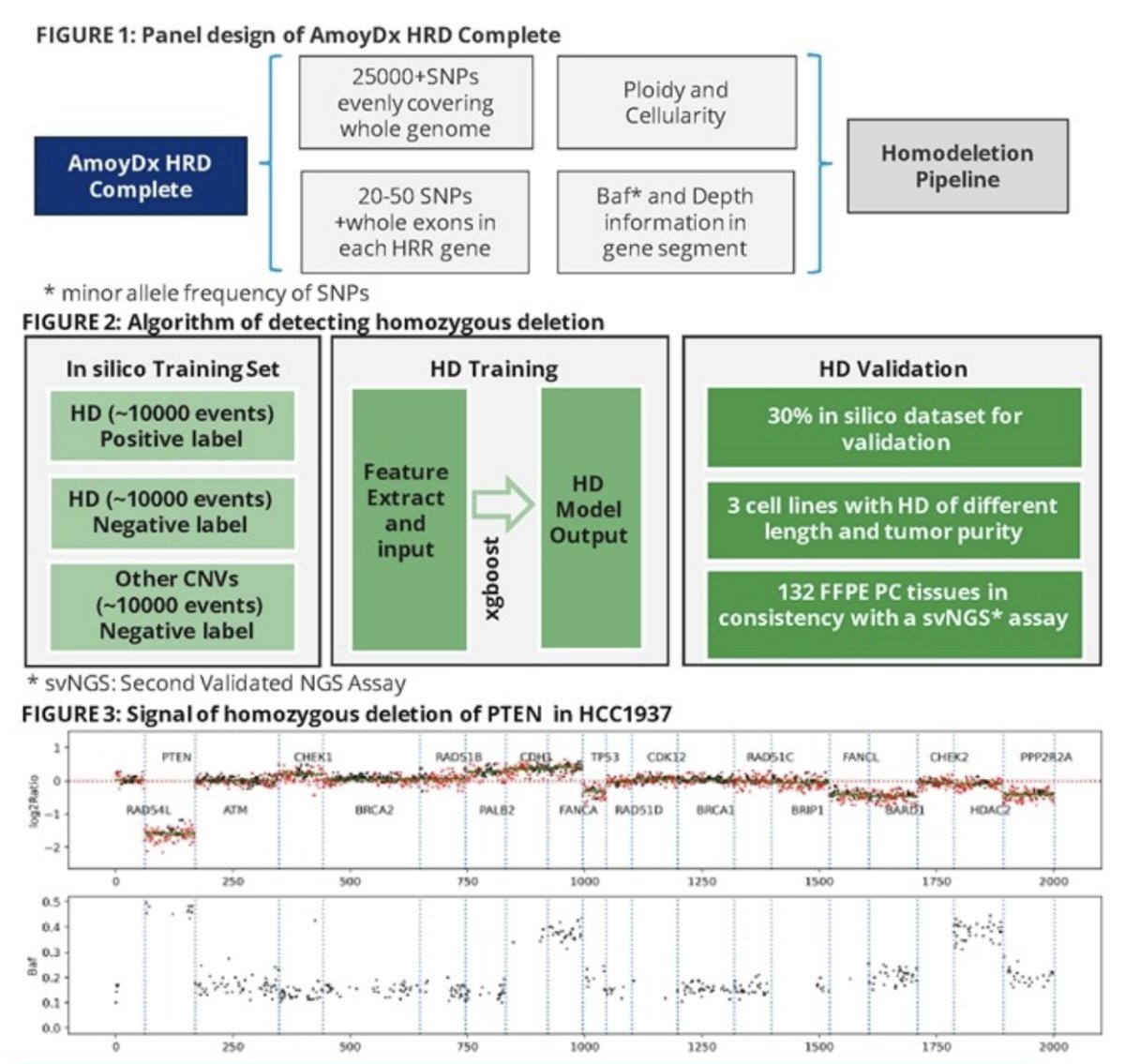
Seven artificial features were calculated based on:
- Genomic segment depth
- Minor allele frequency of single nucleotide polymorphisms
- Tumor cellularity
- Tumor ploidy
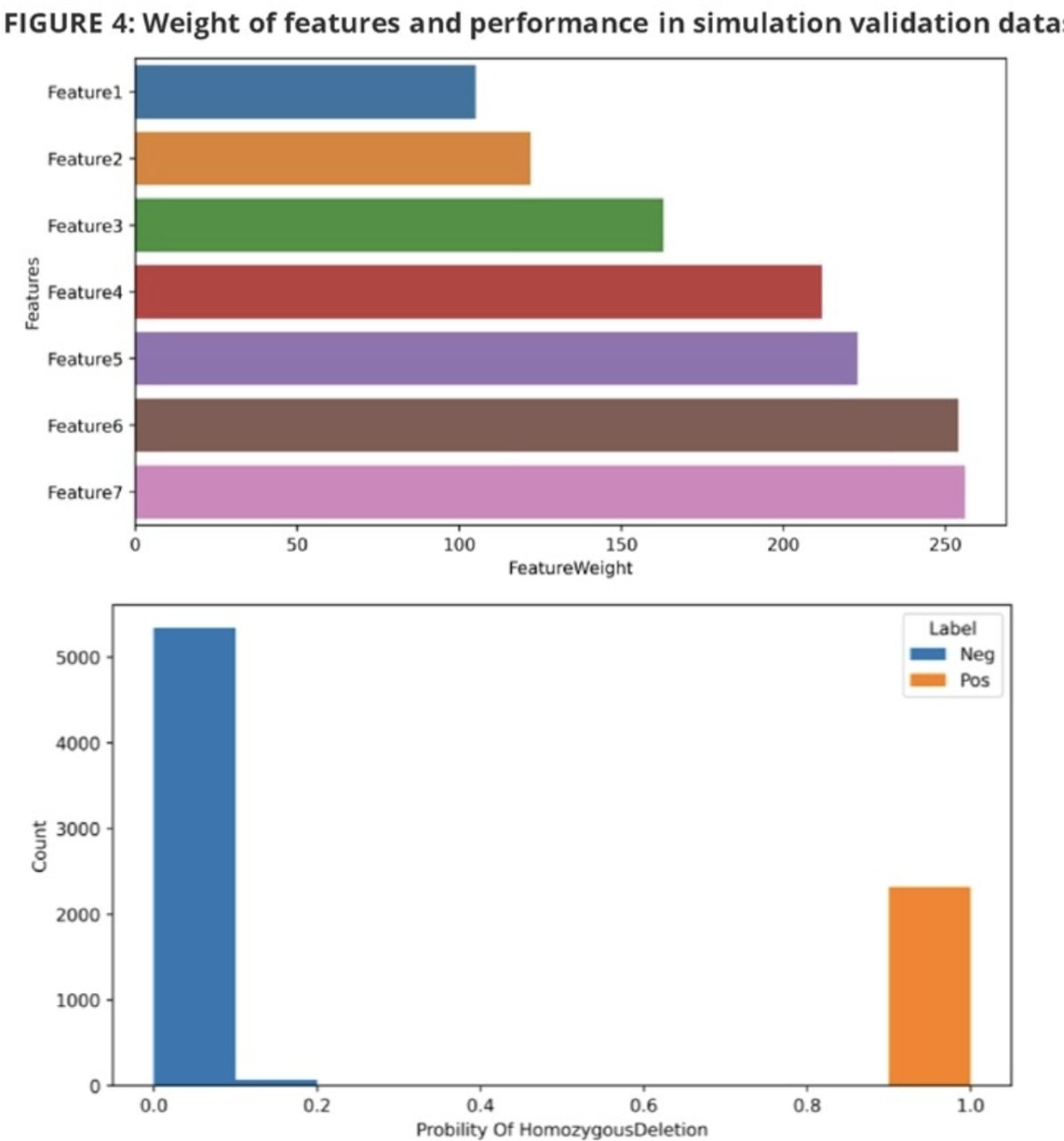
The feature data were fed into the xgboost with 10-StratifiedKFold to generate homozygous deletion recognition models (training to validation datasets ratio 7:3). The analytical performance was assessed using reference cell lines:
- Specificity with wildtype cell lines
- Sensitivity with gradient homozygous deletion-positive cell lines (tumor cellularity: 20%, 30%, 40%, 50%) with varying DNA input (30ng, 50ng, 100ng)
- Reproducibility of intra- and inter-runs
- Anti-interference against hemoglobin, triglycerides, xylene, paclitaxel, and ethanol
The clinical performance was assessed using formalin-fixed paraffin-embedded prostate cancer tissues.
The trained homozygous deletion model achieved 99.8% accuracy in the in-silico validation dataset. Analytical validation demonstrated:
- 100% specificity
- 100% sensitivity at gene level with 30% tumor cellularity and at exon level with 40% tumor cellularity with 100ng DNA input
- 100% reproducibility
- Strong anti-interference capability with all homozygous deletion events detected and no false positives.
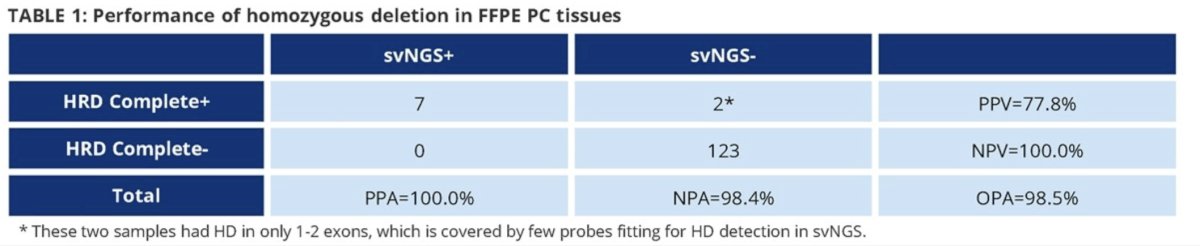
132 formalin-fixed paraffin-embedded prostate cancer tissues showed high consistency with a second validated NGS assay (percent positive agreement: 100%; negative percent agreement: 98.4%; overall percent agreement: 98.5%).

Dr. Wang and colleagues concluded that they were able to successfully develop a machine learning-based model, which was adopted in multiple NGS assays to detect homozygous deletions of HRR genes in prostate cancer tissues. They argued that this model provides a reliable approach to effectively identify prostate cancer patients with HRR homozygous deletions.
Presented by: Jianqing Wang, MD, Xiamen, China
Written by: Rashid K. Sayyid, MD, MSc – Society of Urologic Oncology (SUO) Clinical Fellow at The University of Toronto, @rksayyid on Twitter during the 2023 European Society of Medical Oncology (ESMO) Annual Congress held in Madrid, Spain between October 20th and 24th, 2023
References:- Anscher MS; Chang E; Gao X; Gong Y; Weinstock C; Bloomquist E; Adeniyi O; Charlab R; Zimmerman S; Serlemitsos-Day M; Ning YM; Mayrosh R; Fuller B; Trentacosti AM; Gallagher P; Bijwaard K; Philip R; Ghosh S; Fahnbulleh F; Diggs F; Arora S; Goldberg KB; Tang S; Amiri-Kordestani L (2020) FDA approval summary: Rucaparib for the treatment of patients with deleterious BRCA-mutated metastatic castrate-resistant prostate cancer.
- Abida W, Patnaik A, Campbell D, et al. Rucaparib in Men With Metastatic Castration-Resistant Prostate Cancer Harboring a BRCA1 or BRCA2 Gene Alteration. J Clin Oncol. 2020;38(32):3763-3772.
- Center for Drug Evaluation and Research (2020) FDA approves Olaparib for HRR gene-mutated metastatic castration-resis, U.S. Food and Drug Administration.
- De Bono J, Mateo J, Fizazi K, et al. Olaparib for Metastatic Castration-Resistant Prostate Cancer. N Engl J Med. 2020;382:2091-2102.
- Center for Drug Evaluation and Research (no date b) FDA D.I.S.C.O. burst: Lynparza, U.S. Food and Drug Administration. (2023)
- Clarke NW, Armstrong AJ, Thiery-Vuillemin A, et al. Abiraterone and Olaparib for Metastatic Castration-Resistant Prostate Cancer. NEJM Evid. 2022;1(9).
- ESMO (2023) FDA approves Talazoparib with Enzalutamide for HRR gene-mutated metastatic castration-resistant prostate cancer, ESMO.
- Agarwal N, Azad AA, Carles J, et al. Talazoparib plus enzalutamide in men with first-line metastatic castration-resistant prostate cancer (TALAPRO-2): a randomised, placebo-controlled, phase 3 trial. Lancet. 2023;402(10398):291-303.


