Germline mutations are found in sperm only, whereby the entire organism carries the mutation, and half of the gametes carry the mutation. Somatic mutations only occur in a specific area of the body (ie. in the tumor), are not carried in the gametes, and are not hereditary: 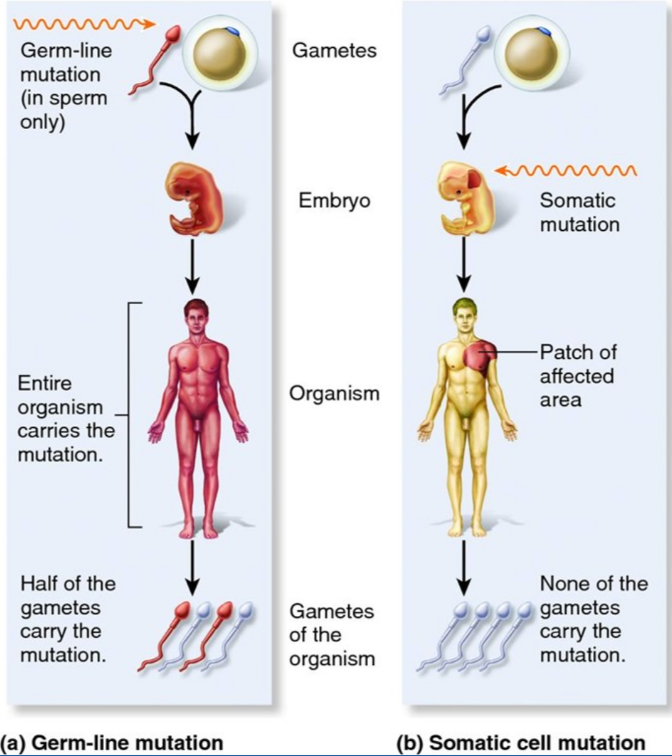
The prostate cancer mutational landscape is dynamic and changes as the disease progresses. Primary prostate cancer is primarily driven by ETS rearrangement, TMPRESS2-ERG, as well as to a lesser degree TP53, and PTEN. Metastatic prostate cancer is primarily driven by AR mutations, PTEN, and TP53. Looking at germline mutations in advanced prostate cancer, Pritchard et al.1 showed that 11.8% of patients with advanced prostate cancer had a germline mutation, primarily DNA repair genes BRCA1, BRCA2, ATM, and CHEK2:

Early data from Mateo et al. suggested that response to olaparib in patients with metastatic prostate cancer was heavily driven by whether the DNA repair defect biomarker was present or not: biomarker positive patients had a median rPFS of 9.8 months vs 2.7 months for the biomarker negative group (p<0.001). Further assessments of the genomic landscape in metastatic castration-resistant prostate cancer (CRPC) found that actionable aberrations are detected in 89% of patients, but Dr. Evans notes that findings something that is truly “actionable” is low, and there is questionable feasibility outside of boutique cancer centers given the cost and technical expertise. Additionally, Dr. Evans’ West Coast Stand Up to Cancer Dream team biopsied 202 patients at their metastatic site with the following biopsy site breakdown:3

Several important findings emerged from this study including that the overall incidence of treatment-emergent small-cell neuroendocrine prostate cancer detection was 17%. Furthermore, AR amplification and protein expression were present in 67% and 75%, respectively, of treatment-emergent small-cell neuroendocrine prostate cancer biopsy specimens. Genomic alterations in the DNA repair pathway were nearly mutually exclusive with treatment-emergent small-cell neuroendocrine prostate cancer differentiation (p = 0.035), and detection of treatment-emergent small-cell neuroendocrine prostate cancer was associated with shortened overall survival among patients with prior AR-targeting therapy for mCRPC (HR 2.02, 95% CI 1.07 to 3.82):

Based on these results, Dr. Evans’ group developed a treatment-emergent small-cell neuroendocrine prostate cancer gene expression signature that had 91% internal accuracy on cross-validation and was enriched for the presence of neural development genes (SEMA3, EPHA7, and TENM3). When assessing overall survival of treatment-emergent small-cell neuroendocrine prostate cancer versus no treatment-emergent small-cell neuroendocrine prostate cancer by gene signature (rather than histology) the median OS was 17.6 months vs 41.7 months (HR 3.00, 95% CI 1.25-7.19):

Dr. Evans’ group published the first whole-genome sequence of metastatic prostate cancer published in 2018 in Cell.4 They found that tandem duplication affects intergenic regulatory loci upstream of AR and MYC, as well as inactivation of CDK12, TP53, and BRCA2 affecting distinct classes of structural variants. Furthermore, they found that the androgen receptor is affected by mutation or structural variation in 85% of patients with mCRPC. Recently, Dr. Evans’ group also published the DNA methylation landscape of advanced prostate cancer in Nature Genetics.5 This study found that whole-genomic bisulfite sequencing paired with deep whole-genome and transcriptome sequencing of 100 castration-resistant prostate metastases. They discovered alterations affecting driver genes that were detectable only with integrated whole-genome approaches. Differential methylation preferentially occurs at somatic mutational hotspots and putative regulatory regions, whereby 22% of tumors exhibited a novel epigenomic subtype associated with hypermethylation and somatic mutations. Finally, they identified intergenic regions where methylation is associated with RNA expression of the oncogenic driver genes AR, MYC, and ERG.
Dr. Evans concluded his presentation with the following take-home messages:
- Germline mutations in mCRPC are predictive of treatment response
- Somatic mutations correlate with histology and survival in mCRPC
- Somatic mutations can be accessed for solid or liquid biopsies
- DNA methylation also drives somatic DNA mutation expression
Presented by: Christopher Evans, MD, FACS, Professor and Chair Department of Urologic Surgery, UC-Davis Medical Center, Sacramento, California
Written by: Zachary Klaassen, MD, MSc – Assistant Professor of Urology, Georgia Cancer Center, Augusta University/Medical College of Georgia, Twitter: @zklaassen_md at the 2020 Société Internationale d'Urologie Virtual Congress (#SIU2020), October 10th - October 11th, 2020
References:
1. Pritchard CC, Mateo J, Walsh MF, et al. Inherited DNA-Repair gene mutations in men with metastatic prostate cancer. N Engl J Med. 2016;375(5):443-453.
2. Mateo J, Carreira S, Sandhu S, et al. DNA-Repair defects and olaparib in metastatic prostate cancer. N Engl J Med. 2015;373(18):1697-1708.
3. Aggarwal R, Huang J, Alumkal JJ, et al. Clinical and Genomic Characterization of Treatment-Emergent Small-Cell Neuroendocrine Prostate Cancer: A Multi-Institutional Prospective Study. J Clin Oncol2018 Aug 20;36(24):2492-2503.
4. Quigley DA, Dang HX, Zaho SG, et al. Genomic Hallmarks and Structural Variation in Metastatic Prostate Cancer. Cell. 2018 Oct 18;175(3):889.
5. Zhao SG, Chen WS, Li H, et al. The DNA methylation landscape of advanced prostate cancer. Nat Genet.2020 Aug;52(8):778-789.


