- Work from the TCGA identified at total of 64 genes with significant evidence of mutation in patients with urothelial carcinoma.
- These include pathways well known to contribute to carcinogenesis including p53 and cell cycle regulation; DNA repair; PI3kinase/AKT; and chromatin modification and regulation.
- Identification of these pathways has led to ongoing work in molecularly-targeted therapeutics.
- Work from the TCGA, in concert with many others, has contributed to our understanding that there are multiple subtypes of muscle-invasive bladder cancer, each of which has differing underlying molecular characteristics, differing intrinsic sensitivity to chemotherapy, and thus differing treatment strategies.
- Confirmation of the importance of smoking in urothelial carcinogenesis.
In this talk, Dr. Lerner began by highlighting insights into the biology of muscle-invasive bladder cancer that were derived from the two published TCGA analyses. Many of these are highlighted in the list above.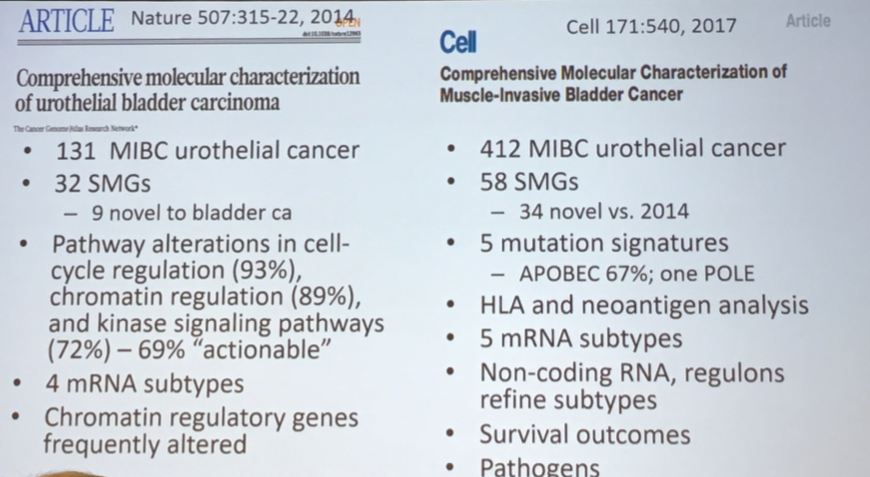
However, an additional key product of the TCGA analysis is a publicly available data repository that allows for significant amplification of the scientific benefit of this work.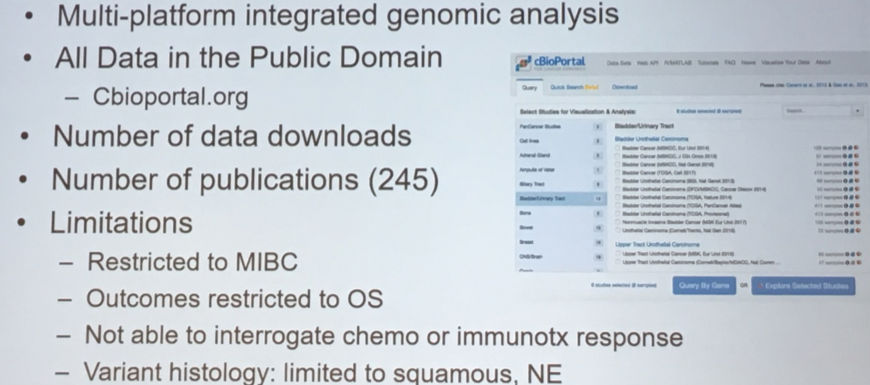
Dr. Lerner highlighted that integration analyses of mutations and amplifications/deletions within the TCGA project has allowed for definitions of bladder cancer subtypes. In addition to allowing classification of bladder tumors, these profiles have important prognostic significance with meaningful differences in survival based on mutational patterns.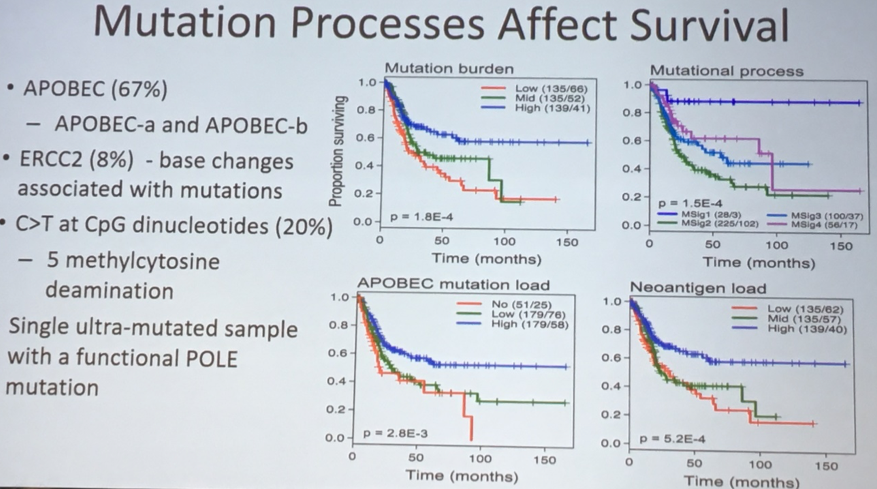
Highlighting the translational applications of the insights derived from the TCGA, Dr. Lerner showed data demonstrating that DNA damage repair signatures can predict response to neoadjuvant chemotherapy.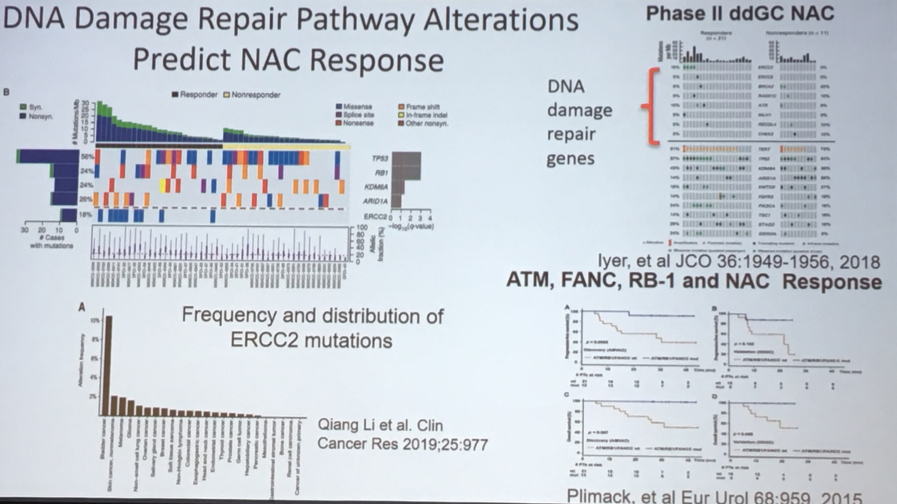
This information has informed the design of the Alliance A031701 trial in which the presence or absence of DNA damage repair alterations guides the allocated treatment.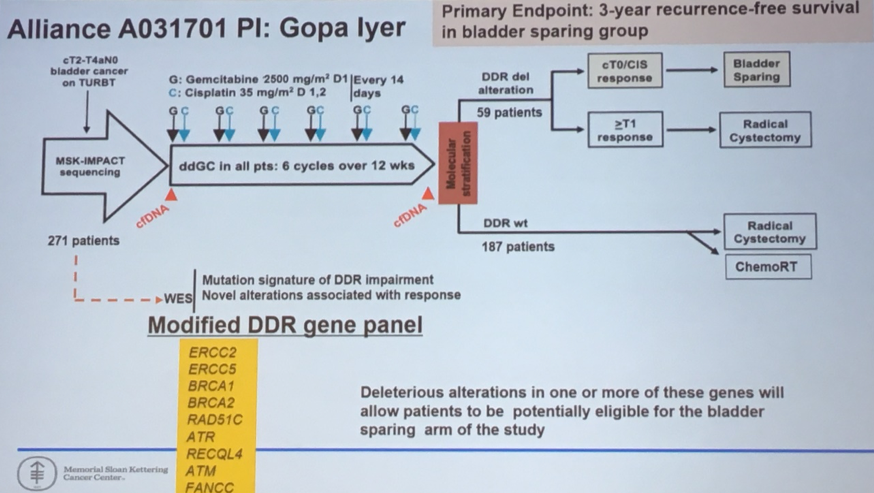
A trial with a similar intent, but using a different molecular profile, is also underway at Fox Chase Cancer Center, ed by Dr. Daniel Geynisman.
Further insights from TCGA-based analyses have also led to the development and approval of novel agents, as well as the recognition of exceptional responders due to underlying mutational signatures. The first of these relates to an insight that the fibroblast growth factor receptor (FGFR) plays an important role in advanced bladder cancer. This led to the development and subsequent approval of erdafitinib. Second, in the context of a negative trial in everolimus, investigators identified a patient with an exceptional response. Whole genome sequencing subsequently identified mutations in TSC1 and 2, underpinning this response.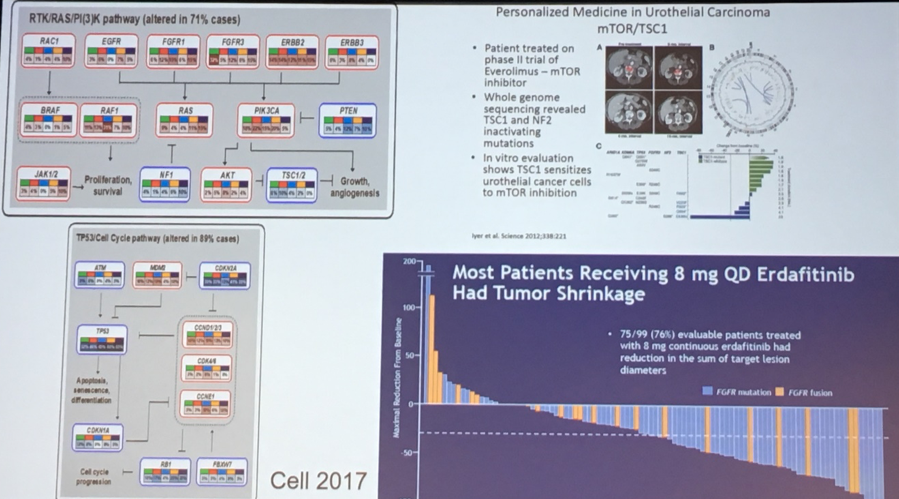
Dr. Lerner then spoke briefly about upper tract urothelial carcinoma, primarily highlighting that, despite the shared histology, UTUC and bladder cancer differ in many molecular characteristics.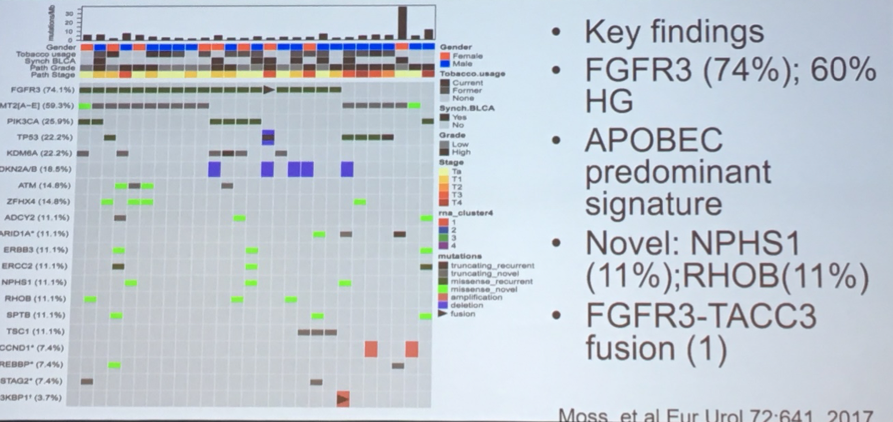
Moving on, Dr. Lerner discussed ongoing work understanding the germline, rather than somatic, genomic landscape of advanced bladder cancer. While the vast majority of patients with urothelial carcinoma demonstrated no germline variants, approximately 14% of patients had such mutations, many of which affected DNA damage repair genes.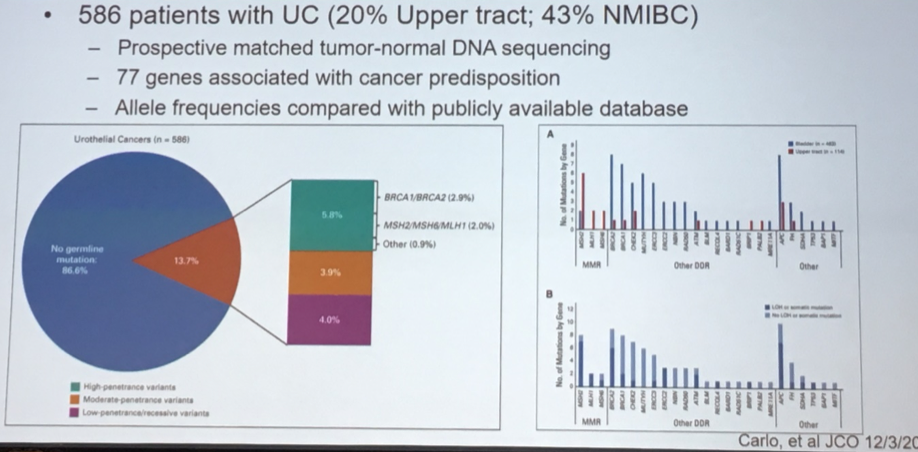
Heat maps of these genetic signatures identified subtypes of urothelial carcinoma with prognostic importance.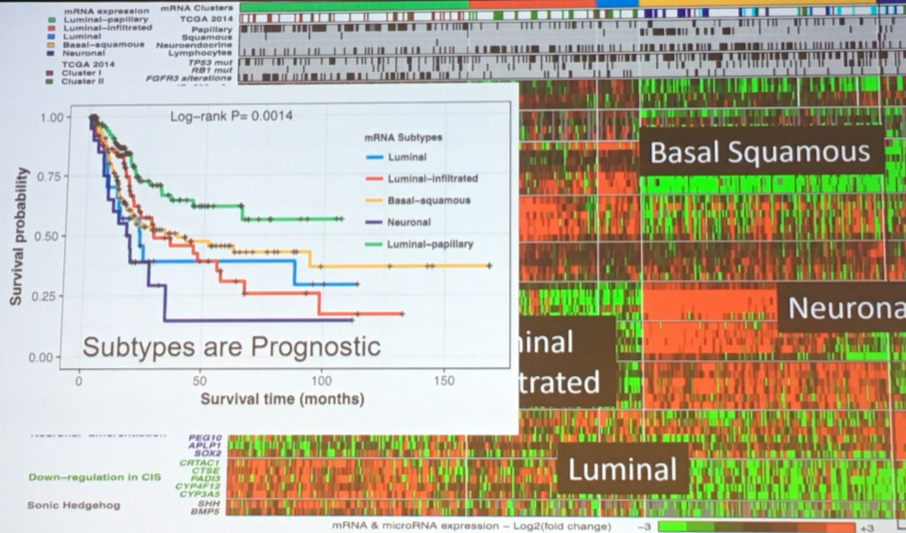
While speculative, these data have led to suggestions of alternative management strategies based on urothelial subtypes, both in the relevance of neoadjuvant therapy and the type of therapy that may be most effective.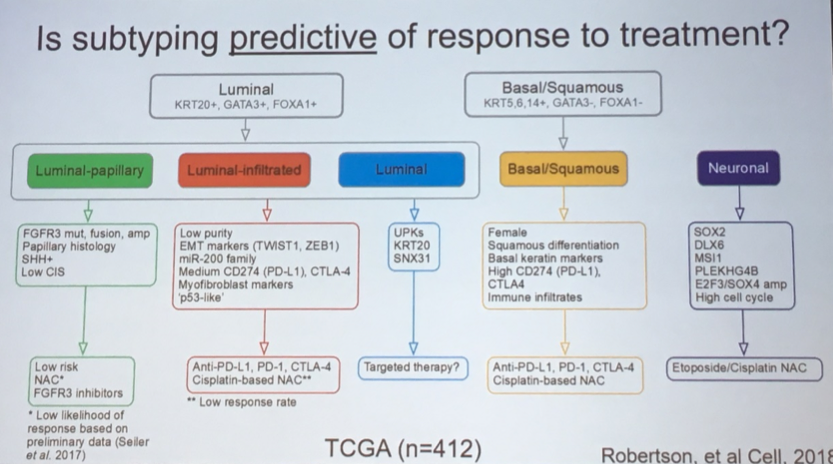
This has further led to the design of a clinical trial in the SWOG cooperative group looking at the use of subtyping to guide management decisions in MIBC.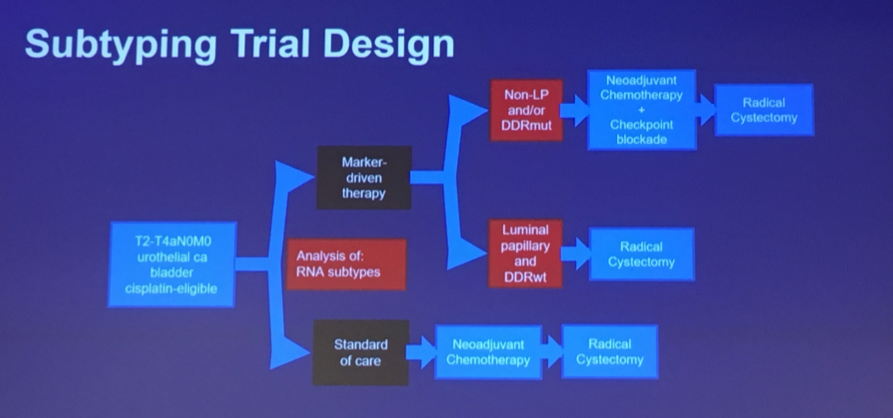
In fact, such a trial is already underway in Europe, under the leadership of Dr. James Catto – the Genotype in Urothelial cancer: Stratified Treatment and Oncological outcomes (GUSTO) trial.
Finally, looking forward, he described opportunities for integration of even more data to provide further insights into bladder cancer carcinogenesis and treatment. In addition to genomic profiles, this would include imaging data and proteomic data. Undertaking this project is the Clinical Proteomic Tumor Analysis Consortium (CPTAC).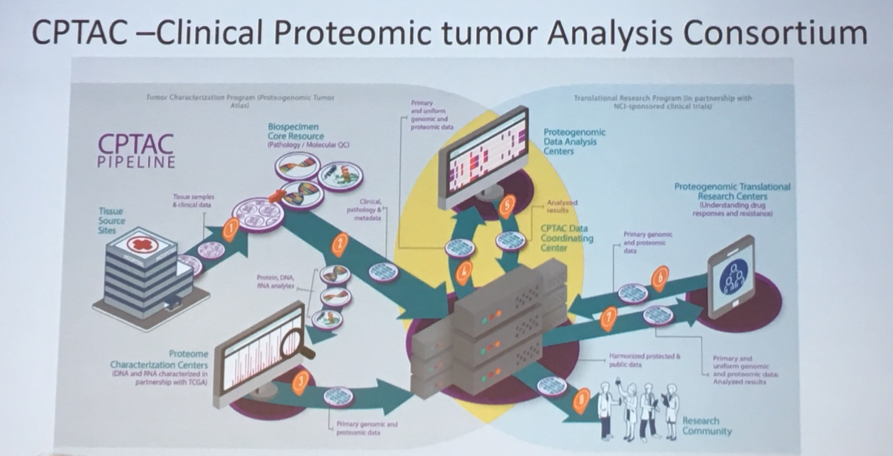
The bladder portion of this study is newly underway and will include newly diagnosed, untreated patients with planned primary cytoreductive surgery, those with new tumors in the context of existing or concurrent tumors, as well as recurrent, metastatic or second primary tumors. Patients with prior cancers or systemic cancer therapy in the preceding 12 months are excluded. Blood and urine specimens will be collected for analysis in addition to standard clinicopathologic data.
Presented by: Seth Paul Lerner, M.D., FACS, Professor of Urology, Baylor College of Medicine, Houston, TX, US, Beth and Dave Swalm Chair in Urologic Oncology, Director of Urologic Oncology, Director of the Multidisciplinary Bladder Cancer Program
Written by: Christopher J.D. Wallis, MD, PhD, FRCSC, @WallisCJD on Twitter, at the 20th Annual Meeting of the Society of Urologic Oncology (SUO), December 4 - 6, 2019, Washington, DC
References:
- Cancer Genome Atlas Research N. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014; 507(7492): 315-22.
- Robertson AG, Kim J, Al-Ahmadie H, et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell 2018; 174(4): 1033.


