(UroToday.com) The Society of Urologic Oncology (SUO) annual winter meeting included a health services research session and a presentation by Dr. Anobel Odisho discussing clinical informatic strategies to accelerate research in urologic oncology.
Dr. Odisho started by noting that aspects of quality improvement, research, and care delivery were historically separate entities but recently have many overlapping aspects:

Dr. Odisho emphasized that this is not a big data talk, but rather informatics is the science of how to use data, information, and knowledge to improve human health and the delivery of health-care services: ie. human-computer interaction meets clinical workflows. There are four different ways to leverage data, as highlighted by Dr. Odisho:
- Use existing data
- This is routinely collected as part of clinical care, it is cheap and readily available
- For this type of data, it is important to understand structured versus unstructured data
- This data is rarely useful in its raw form and needs clinical context
- Modify the electronic health record (EHR)
- The EHR is constantly changing
- It is expensive upfront but has a widespread, long-term impact
- There is a need to change human workflows in parallel
- Created derived data
- Extract meaning from unstructured data
- Use logic and rule to determine events and outcomes
- Enter manual data
- The highest cost over the long-term
- It can arbitrate data conflicts
- It is best when judgment is required or analog records
Using Existing Data
It is important when using existing data to ascertain what is structured and what is unstructured data. Structured data includes demographics, codes (ie. CPT, procedure, ICD), labs, orders, events, metadata, digital dust, flowsheets, smart date elements, and patient-reported outcomes. Unstructured data includes clinical notes, radiology images, and reports, pathology images and reports, patient-reported outcomes, photos, videos, email, phone calls, secure messages, and scanned documents.
Modify the EHR
Typically, ICD-10 codes are haphazardly used and are historically poor at capturing diagnoses. Improved granularity is obtained by adding SNOMED-CT codes such as code 461511000124101 for biochemically recurrent prostate cancer rather than C61 for prostate cancer. Using data entry embedded into a workflow is fast and ‘smart data elements' are stored in databases automatically. An example of this is embedding smart data elements for adding nephrometry scores to the workflow. Furthermore, pre-populated operative notes can also have data elements that pre-populate post-procedure notes.
Create Derived Data
A Urology Data Warehouse includes a cloud-based SQL Server directly linked to the EHR database, which is continuously updated (837 nightly scheduled data tasks and counting). A model for this workflow is as follows:
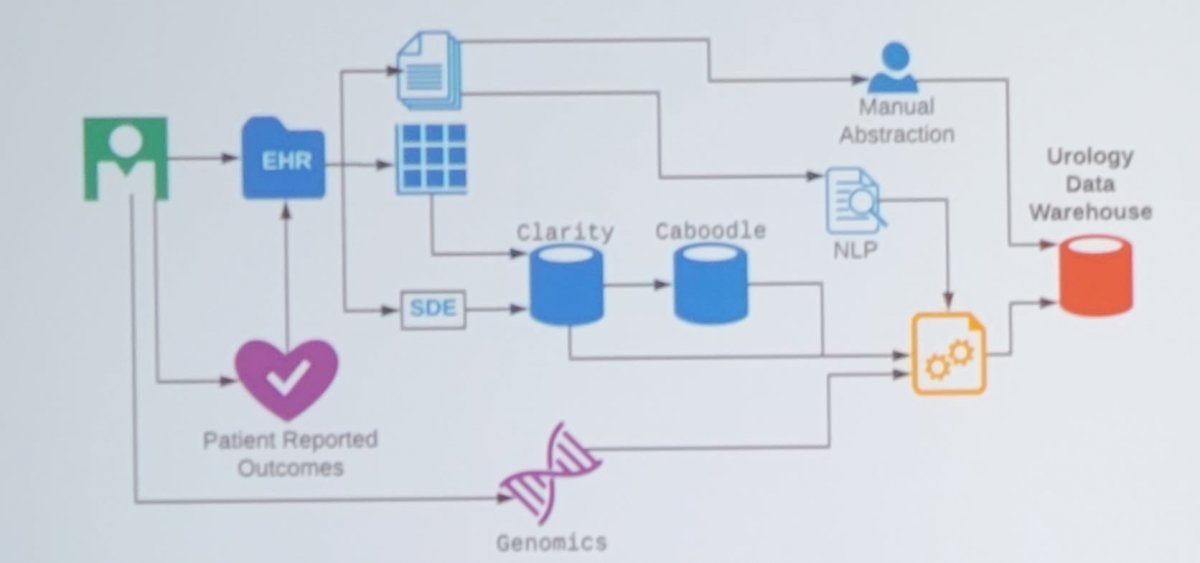
Dr. Odisho notes that rules-based natural language processing (NLP) is used to extract and parse reports from the EHR nightly. At UCSF, >9,700 prostate biopsy reports and >16,500 MRI reports have been parsed using regular expressions and pattern matching. Dr. Odisho and colleagues published their NLP system in 2019, assessing 523 patients with NLP-extracted data, 319 with structured data elements data, and 555 with manually abstracted data1. They found that for Gleason scores, NLP and clinician structured data element accuracy was 95.6% and 95.8%, respectively, compared with manual abstraction, with concordance of 0.93 (95% CI, 0.89 to 0.98). For margin status, extracapsular extension, and seminal vesicle invasion, stage, and lymph node status, NLP accuracy was 94.8% to 100%, structured data element accuracy was 87.7% to 100%, and concordance between NLP and structured data element ranged from 0.92 to 1.0. As follows is the comparison of accuracy and concordance among the three data abstraction methods:

Dr. Odisho emphasized that it is important to create a context for data. In the UCSF database, there are 236,233 PSA values, but alone they do not have any meaning. To catalog a PSA at diagnosis, this needs a date of diagnosis; for a first PSA after surgery, this needs a date of surgery; to adjust for a patient taking 5-alpha reductase inhibitor, this needs a medication list. To do this, one must use a series of continually re-running logical rules to create contextual values.
Data validation for derived data is as follows:

Enter Manual Data
Dr. Odisho notes that some values are otherwise inaccessible, such as outside the institution results. Manually entering data allows manual verification of elements that are flagged for review, and whenever possible these values should be entered directly into the EHR. Importantly, there should be a separate interface for data entry and validation.
There are several examples of how this data may be used. For example, this includes enrollment for SMS-based prostate biopsy patient digital engagement and biobank specimen inventory management. Dr. Odisho concluded his presentation by summarizing that we need to (i) plan what data you need, (ii) build into existing workflows, (iii) invest money in the long-term, and (iv) start small.
Presented by: Anobel Y. Odisho, MD, MPH, Departments of Urology, Epidemiology & Biostatistics, University of California San Francisco, San Francisco, CA
Written by: Zachary Klaassen, MD, MSc – Urologic Oncologist, Assistant Professor of Urology, Georgia Cancer Center, Augusta University/Medical College of Georgia, @zklaassen_md on Twitter during the 2021 Society of Urologic Oncology (SUO) Winter Annual Meeting, Orlando, FL, Wed, Dec 1 – Fri, Dec 3, 2021.
References:
- Odisho AY, Bridge M, Webb M, et al. Automating the Capture of Structured Pathology Data for Prostate Cancer Clinical Care and Research. JCO Clin Cancer Info. 2019. Online July 17, 2019


