(UroToday.com) The Society of Urologic Oncology (SUO) 2021 annual meeting in Orlando, FL hosted an overview of emergent biomarkers to identify patients with high-risk non-metastatic renal cell carcinoma (RCC) presented by Dr. Brandon Manley, MD. Dr. Manley began his talk by discussing methylation profiles in clear cell RCC (ccRCC). Wei et al. used genome-wide CpG methylation profiling, a LASSO model to develop a five-CpG-based assay for ccRCC prognosis. The five CPG-based classifiers were validated in three independent sets from China, the United States, and the Cancer Genome Atlas data set, predicting overall survival of ccRCC patients with a HR of 2.96-4.82, p<0.001. The five-CpG-based classifier categorizes patients into high and low-risk groups and the methylation profile correlates with expression of 5 genes: PITX1, FOXE3, TWF2, EHBO1L1, and RIN1.1

When the methylation characteristics of short-term survivors (STS) and long-term survivors (LTS) were compared, there appeared to be a trend toward hypermethylation in STS tumors, although this was not statistically significant.2

Nuzzo et al. were recently able to utilize cell-free methylated DNA immunoprecipitation and high-throughput sequencing to accurately classify patients across all stages of RCC in plasma (Area Under Curve [AUC]=0.99) and demonstrate the validity of this assay to identify patients with RCC using urine cell-free DNA (AUC=0.86). The image below depicts differentially methylated regions in cell free DNA (DNA) between RCC and control samples,with the left panel depicting methylation gain and the right panel depicting methylation loss.

Dr. Manley likened this assay to a barcode that can be used to potentially discern each individual patient’s prognosis, optimal treatment, etc.
Next, Dr. Manley addressed the issue of spatial immune cell patterns in RCC. Immune infiltrate is measured as: classically cell density / % Cells positive. In work from the Moffitt Cancer Center, 122 patients with RCC (97 clear cell, 17 papillary, five chromophobe, 3 other) across all stages (~30% stage IV), a multiplex immunofluorescence technique was used to assess lymphoid (CD3, CD8, FOXP3, T-bet) and myeloid (CD68, CD163, CD206, CD20) panels. The investigators focused on the tumor/surrounding environment interface, evaluating tumor/environment affinity for immune cells, visualizing the “tug-of-war” of these cells across the two interfaces. The question becomes whether these cells are clustered within the tumor or the stroma. Tumor/CD163 interface clustering was evaluated across the various RCC stages as depicted in the image below.
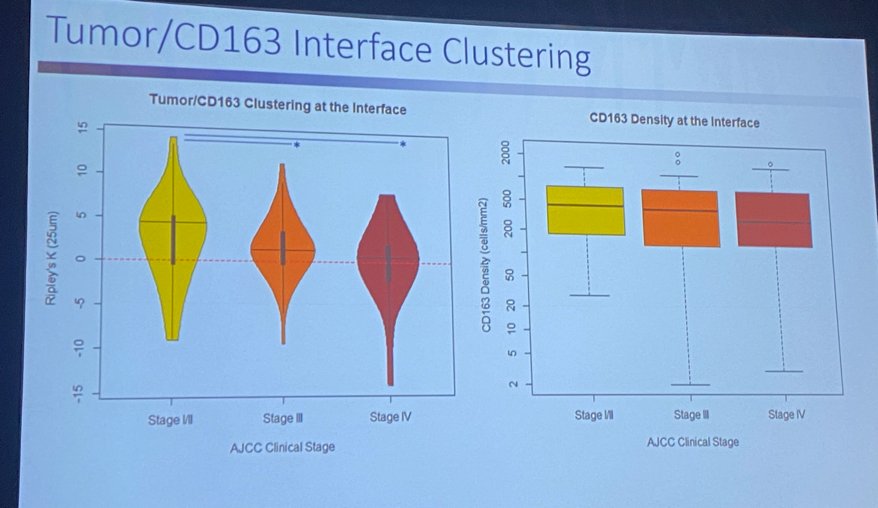
CD163, a marker of perivascular macrophages, was noted to cluster differentially based on the stage of the tumor. As illustrated in the image below, CD163 clustered within the tumor in earlier stages and in the stroma with advanced tumor stages. This “march of the macrophages” was dubbed a tumor Quick Response (QR) code for a patient by Dr. Manley.

This tumor/CD163 clustering can be used as a gene signature to predict overall survival. With “high clustering” a favorable prognostic factor in both the Moffitt and TCGA cohorts.

Dr. Manley went on to describe current efforts by his team to quantify T- and B-cell immune receptor distribution diversity in ccRCC, whereby tumor is biopsied, undergoes bulk RNA sequencing, CDR3 is recovered in sequencing and used to calculate receptor diversity.


The degree of T- and B-cell diversity in ccRCC tunmor is clearly associated with clinical outcomes, whereby high diversity tumors were associated with superior probability of survival.

Finally, Dr. Manley went on to discuss the importance of RNA splicing in ccRCC. Aberrant splicing is only seen in specific states (e.g. cancer). Splicing events tend to be consistent (i.e. a hotspot). They create less “noise” compared to gene expression. Defining splicing or neojunction is method dependent, but the biologic or clinical relevance remains unclear. EGFR appears to be one of the most common splice variants or hotspots seen in RCC. These splice variants have a similar frequency of events across multiple cohorts. Notably, a risk score based on the frequency of these splice variants can be used to predict overall and cancer-specific survival.

Presented by: Brandon Manley, MD, Assistant Professor, Genitourinary Oncology, Moffitt Cancer Center, University of South Florida, Tampa, FL
Written by: Rashid Sayyid, MD, MSc – Urology Chief Resident, Augusta University/Medical College of Georgia, @rksayyid on Twitter during the 2021 Society of Urologic Oncology (SUO) Winter Annual Meeting, Orlando, FL, Wed, Dec 1 – Fri, Dec 3, 2021.
References:
- Wei JH, Haddad A, Wu K, et al. A CpG-methylation-based assay to predict survival in clear cell renal cell carcinoma. Nature Comm. 2015 Oct;6;8699.
- El Khoury LY, Fu S, Hlady RA, et al. Identification of DNA methylation signatures associated with poor outcome in lower-risk Stage, Size, Grade and Necrosis (SSIGN) score clear cell renal cell cancer. Clin Epigenetics. 2021 Jan; 13(1):12.
- Nuzzo PV, Berchuck JE, Korthauer K. Detection of renal cell carcinoma using plasma and urine cell-free DNA methylomes. Nat Med. 2020 Jul;26(7):1041-1043.


