Across all aspects of daily life, artificial intelligence more than ever is harnessing the power of machine learning to automate and improve the efficiency of tasks. This is also true in medicine and, in particular, in the field of prostate cancer. The concept of artificial intelligence began in the 1950s with the prime objective of emulating the cognitive capabilities of human beings in machines. More specifically, artificial intelligence is the ability of a machine to independently replicate intellectual processes typical of human cognition. Machine learning comprises algorithms that parse data, learn from that data, and then apply what they have learned to make informed decisions. Deep learning is a form of machine learning that is inspired by the structure of the human brain and is particularly effective in feature detection:1

This Center of Excellence article will highlight advances in the field of artificial intelligence with applications for the diagnosis and grading of prostate cancer, the use and interpretation of multiparametric MRI, and active surveillance in low risk prostate cancer.
Artificial Intelligence for the Diagnosis and Grading of Prostate Cancer
The Gleason grading system has been the most reliable tool for the prognosis of localized prostate cancer since its development. However, its clinical application may be limited by interobserver variability in grading and quantification, which has negative consequences for risk assessment and clinical management of prostate cancer. As such, several studies have examined the impact of an artificial intelligence-assisted approach to prostate cancer grading and quantification. Marginean and colleagues2 developed an artificial intelligence algorithm based on machine learning and convolutional neural networks, as a tool for improved standardization in Gleason grading in prostate cancer biopsies. This study included 698 prostate biopsy sections from 174 patients that were used for training, with a final algorithm tested on 37 biopsy sections from 21 patients, with digitized slide images from two different scanners. Overall, this algorithm had a high accuracy in detecting cancer areas with 100% sensitivity and 68% specificity. Compared to pathologists, the algorithm also performed well in detecting cancer areas with an intraclass correlation coefficient of 0.99 and assigning the Gleason patterns correctly (Gleason patterns 3 and 4 ICC: 0.96 and 0.94, respectively; Gleason pattern 5 ICC: 0.82).
Huang et al.3 have also reported their results of a single center assessment examining the impact of an artificial intelligence-assisted approach to prostate cancer grading and quantification. This study included 589 men with biopsy-confirmed prostate cancer who received care from January 1, 2005, through February 28, 2017. A total of 1,000 biopsy slides were selected and scanned to create digital whole-slide images. These were subsequently used to develop and validate a deep convolutional neural network-based artificial intelligence-powered platform. These whole-slide images were then divided into a training set (n = 838) and validation set (n = 162). As follows is an example of the artificial intelligence-power platform, with the left panel showing results of quantitative Gleason grading and quantification of prostate cancer volume, while the right panel shows biopsy cores with annotations independently made by artificial intelligence:
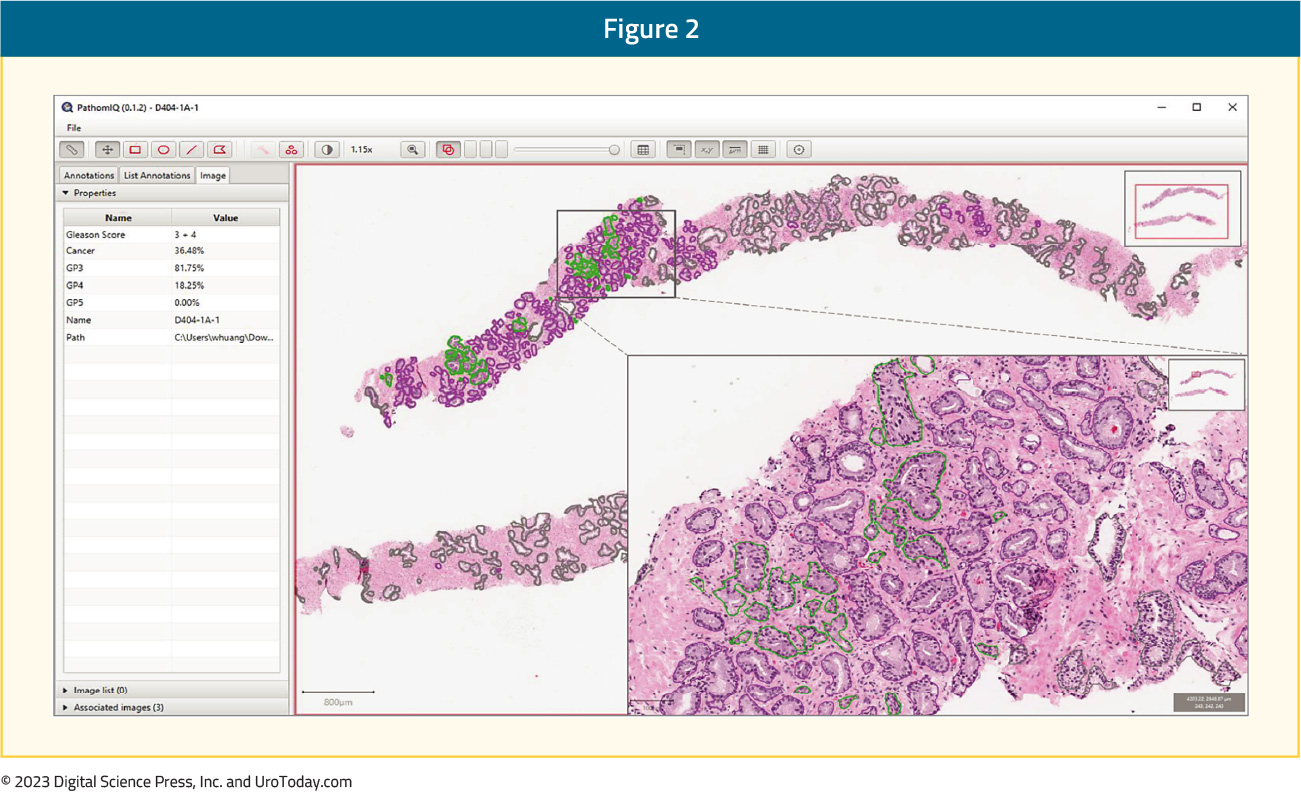
Ultimately, the artificial intelligence system was able to distinguish prostate cancer from benign prostatic epithelium and stroma with high accuracy at the patch-pixel level, with an area under the receiver operating characteristic curve of 0.92 (95% CI, 0.88-0.95). Furthermore, the artificial intelligence system achieved an almost perfect agreement with the training pathologist in detecting prostate cancer at the patch-pixel level (weighted κ = 0.97) and in grading prostate cancer at the slide level (weighted κ = 0.98). The use of the artificial intelligence-assisted method was also associated with significant improvements in the concordance of prostate cancer grading and quantification between three pathologists and significantly higher weighted κ values for all pathologists compared with the manual method.
More recently, Bulten et al.4 published results of the PANDA challenge, the largest histopathology competition to date, which was comprised of 1,290 developers to catalyze development of reproducible artificial intelligence algorithms for Gleason grading using 10,616 digitized prostate biopsies. As follows is an overview of the PANDA challenge and the study setup:

This study validated that a diverse set of submitted algorithms reached pathologist-level performance on independent cross-continental cohorts, fully blinded to the algorithm developers. Using the United States and European external validation sets, the algorithms achieved concordance of 0.862 (quadratically weighted κ, 95% CI, 0.840-0.884) and 0.868 (quadratically weighted κ, 95% CI, 0.835-0.900) with expert uropathologists.
Previous work using machine learning has also assessed algorithms for predicting adverse outcomes after radical prostatectomy using whole-slide images of prostate biopsies with Grade Group 2 or 3 disease. Paulson and colleagues5 retrospectively reviewed prostate biopsies of patients who had corresponding radical prostatectomy data, Grade Group 2 or 3 disease in one or more cores, and no biopsies with higher than Grade Group 3 disease. The machine learning pipeline in this study had three phases: image preprocessing, feature extraction, and adverse outcome prediction. This data set included 361 whole-slide images from 107 patients, including 56 with adverse pathology at radical prostatectomy. The area under the receiver operating characteristic curves for the machine learning classification were 0.72 (95% CI, 0.62 to 0.81), 0.65 (95% CI, 0.53 to 0.79), and 0.89 (95% CI, 0.79 to 1.00) for the entire cohort, Grade Group 2 and Grade Group 3 patients, respectively, and had a similar discriminatory performance to that of the CAPRA clinical risk assessment.
Current artificial intelligence systems have shown that they can perform Gleason grading on par with expert uropathologists and that they can assist pathologists to achieve higher agreement with consensus grading. Current development also indicates that artificial intelligence systems may be trained using long-term outcomes to achieve higher agreement with prognosis:6
As has been highlighted, one of the values of artificial intelligence based, or informed, histopathologic assessment is the potential for increased consistency. It has been recognized for years that sub-specialization within medicine provides benefits. In the field of pathology, sub-specialized genitourinary pathologists are often relied upon as the ground truth. In very recently published work, Jung and colleagues showed that general pathologists, when assisted by an AI algorithm, DeepDx, achieved significantly improved concordance with the genitourinary pathologists’ assessment.7 Thus, widespread adoption of this approach may help to address disparities in the quality of care available to patients living in small communities and more rural areas where sub-specialty expertise is not readily available.
Artificial intelligence for the diagnosis and grading of prostate cancer can reliably detect prostate cancer and quantify the Gleason patterns in core needle biopsies, with similar accuracy as pathologists. Furthermore, these results are reproducible on images from different scanners with a low level of intraobserver variability. Artificial intelligence-powered platforms may potentially transform histopathological evaluation and improve risk stratification and clinical management of prostate cancer.
Artificial Intelligence and Prostate Multiparametric MRI
One of the earliest and most impactful areas for artificial intelligence has been in the field of imaging. Men suspected of harboring prostate cancer increasingly undergo multiparametric MRI and MRI-guided biopsy, and thus the potential of multiparametric MRI coupled to artificial intelligence methods to detect and classify prostate cancer before decision-making has been investigated.8 Mehralivand et al.9 previously reported results of a multicenter multi-reader evaluation of an artificial intelligence-based attention mapping system for the detection of prostate cancer with multiparametric MRI. In this study, multiparametric MRIs from five institutions were included and were evaluated by nine readers. In the first round, readers evaluated multiparametric MRI studies using the PI-RADS v2, and after 4 weeks images were again presented to readers along with the artificial intelligence-based detection system output. Readers accepted or rejected lesions within four artificial intelligence -generated attention map boxes. The performances of readers using the multiparametric MRI-only and artificial intelligence-assisted approaches were compared. Overall, there were 152 case patients and 84 control patients with 274 pathologically proven cancer lesions. The lesion-based area under the curve was 74.9% for MRI and 77.5% for artificial intelligence with no significant difference (p = 0.095). However, the sensitivity for overall detection of cancer lesions was higher for artificial intelligence than for multiparametric MRI but did not reach statistical significance (57.4% vs 53.6%, p = 0.073). For transition zone lesions, sensitivity was higher for artificial intelligence than for MRI (61.8% vs 50.8%, p = 0.001) as highlighted in the following figure:

Furthermore, Hiremath et al.10 recently published an analysis assessing an integrated nomogram combining deep learning, PI-RADS scoring, and clinical variables for identification of clinically significant prostate cancer on biparametric MRI. Among 592 patients (823 lesions) with prostate cancer who underwent multiparametric MRI the integrated clinical nomogram yielded an area under the curve of 0.81 (95% CI 0.76 - 0.85) for identification of clinically significant prostate cancer in the validation data set, significantly improving performance over the deep learning-based imaging predictor (0.74, 95% CI 0.69 - 0.80) and PI-RADS score (0.76 95% CI 0.71 - 0.81) nomograms. In a subset of patients who had undergone radical prostatectomy (n=81), the integrated clinical nomogram resulted in a significant separation in the survival curves between patients with and without biochemical recurrence (HR 5.92, 95% CI 2.34 - 15.00), whereas the deep learning-based imaging predictor (1.22, 95% CI 0.54 - 2.79) and PI-RADS score nomograms did not (1.30, 95% CI 0.62 - 2.71). Hosseinzadeh et al.11 assessed deep learning-assisted prostate cancer detection on bi-parametric MRI utilizing 2,734 consecutive biopsy-naïve men with an elevated PSA who underwent multiparametric MRI that were first scored with PI-RADS v2 followed by development of deep-learning framework designed and trained to predict PI-RADS ≥ 4 lesions from bi-parametric MRI. Overall, the deep learning sensitivity for detecting PI-RADS ≥ 4 lesions was 87% (95% CI 82 - 91) at an average of one false positive per patient, and an area under the curve of 0.88 (95% CI 0.84 - 0.91). The deep learning sensitivity for the detection of Gleason > 6 lesions was 85% (95% CI 77 - 83) at an average of one false positive compared to 91% (95% CI 84 - 96) at an average of 0.3 false positives for a consensus panel of expert radiologists.
Several phase 3 trials have shown the utility of MRI-guided targeted biopsy, and this may also be another avenue for which artificial intelligence may be of use. Recently, ElKarami et al.8 published results of their study assessing the ability of machine learning methods to predict upgrading of Gleason score on confirmatory MRI-guided targeted biopsy of the prostate in 592 men who may be candidates for active surveillance. Upgrading to significant prostate cancer on MRI-guided targeted biopsy was defined as upgrading to Gleason 3+4 and Gleason 4+3, for which machine learning classifiers were applied on both upgrading definitions. For upgrading prediction, the AdaBoost model was highly predictive of upgrading to Gleason 3+4 (area under the curve 0.952), while for prediction of upgrading to Gleason 4+3, the Random Forest model had a lower but excellent prediction performance (area under the curve 0.947).
Lesion detection and classification performance metrics are promising but require prospective implementation and external validation for clinical significance. Ultimately, for an algorithm to be implemented into radiological workflows and to be clinically applicable, it must be trained with a ground truth labeling that reflects histopathological information for the entire prostate, and it must be externally validated.
Artificial Intelligence in Active Surveillance
Active surveillance is guideline-recommended management for patients with low-grade low volume prostate cancer, with multiple longitudinal institutional studies demonstrating its safety and feasibility, while concomitantly avoiding unnecessary side-effects of primary therapy. However, a robust method to incorporate "live" updates of progression risk during follow-up has. to date, been lacking. To address this unmet need, Lee et al.12 developed a deep learning-based individualized longitudinal survival model that learns data-driven distribution of time-to-event outcomes. To further refine outputs, they used a reinforcement learning approach for temporal predictive clustering to discover groups with similar time-to-event outcomes to support clinical utility. Subsequently, these methods were applied to data from 585 men on active surveillance with comprehensive follow-up at a median of 4.4 years. As follows is an illustration of the deep learning-based individualized longitudinal survival model and its use in prediction modeling:

Both Cox and the deep learning-based individualized longitudinal survival models including only baseline variables showed comparable C-indices, but with the deep learning-based individualized longitudinal survival model performance improved with additional follow-up data. With 3 years of data collection and 3 years follow-up, the deep learning-based individualized longitudinal survival model had a C-index of 0.79 (±0.11) compared to 0.70 (±0.15) for landmarking Cox and 0.67 (±0.09) for baseline Cox only. Additionally, the reinforcement learning approach for temporal predictive clustering method further discovered four distinct outcome-related temporal clusters with distinct progression trajectories. Those in the lowest risk cluster had negligible progression risk while those in the highest cluster had a 50% risk of progression by 5 years:

Nayan and colleagues also have assessed whether a machine learning approach could improve prediction of progression on active surveillance.13 In their study, they included 790 patients diagnosed with very-low or low-risk prostate cancer between 1997 and 2016 and managed with active surveillance at Massachusetts General Hospital. In the training set, they trained a traditional logistic regression classifier, and alternate machine learning classifiers (support vector machine, random forest, a fully connected artificial neural network, and machine learning-logistic regression) to predict grade-progression. Over a median follow-up of 6.29 years, 234 men developed grade-progression. As follows are the F1 scores for the various classifiers:

Importantly, all alternate machine learning models had a significantly higher F1 score than the traditional logistic regression model (all p <0.001).
Conclusions
Over the last several years there has been increasing interest in the use of artificial intelligence and incorporation into the diagnosis and management of prostate cancer. Artificial intelligence and machine learning show great promise for the diagnosis and Gleason grading of prostate cancer from digital histopathology images. Although somewhat in its infancy, artificial intelligence algorithms are being reported for multiparametric MRIs and MRI-guided targeted biopsies, and for evaluating “real-time” grade progression for men on active surveillance for low-grade prostate cancer.
Written by:
- Zachary Klaassen, MD MSc, Medical College of Georgia, Augusta, Georgia, USA
- Rashid K. Sayyid, MD MSc, University of Toronto, Toronto, ON


