RIO DE JANEIRO, BRAZIL (UroToday.com) - Presented by Pearce M,1 Hilt E,1 Zilliox M,1 Rosenfeld A,1 White K,1 Tulke M,1 Fok C,1 Mueller E,1 Schreckenberger P,1 Brubaker L,1 Gai X,1 and Wolfe A1 at the International Continence Society (ICS) 2014 Annual Meeting - October 20 - 24, 2014 - Rio de Janeiro, Brazil
ABSTRACT:
Hypothesis/Aims of Study
The presence or absence of lower urinary tract symptoms.[1, 2] This knowledge opens opportunities to explore potential bacterial causes for lower urinary tract symptoms, including urgency urinary incontinence (UUI). In this study, we aimed to compare the urinary microbiome of women with UUI to the microbiome of asymptomatic controls.
Study Design, Materials, and Methods
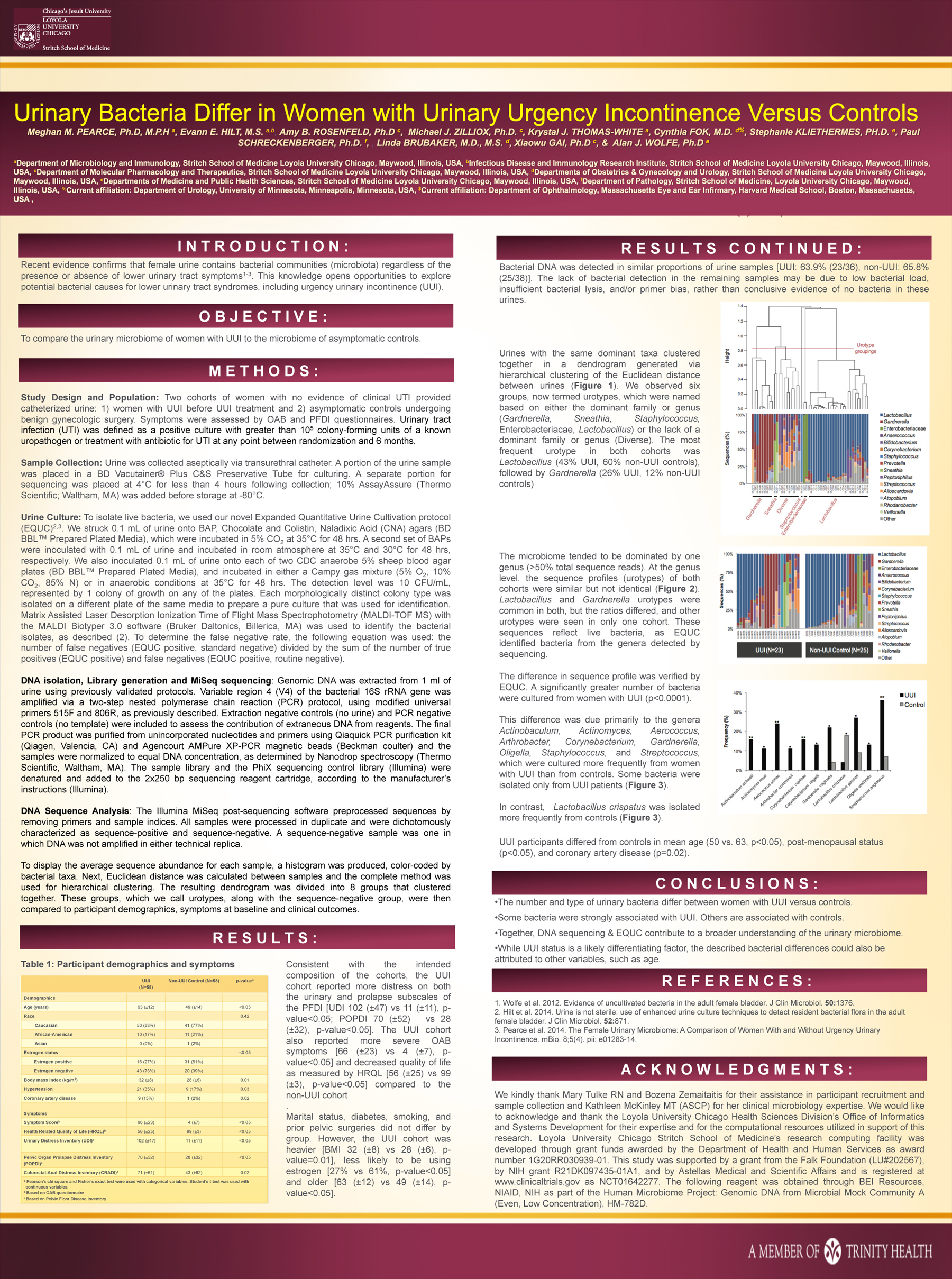 Two cohorts of women with no evidence of clinical UTI provided catheterized urine: 1) women with UUI before UUI treatment and 2) asymptomatic controls undergoing benign gynecologic surgery. Symptoms were assessed by OAB and PFDI questionnaires. To characterize the microbiome, we screened urine for bacterial DNA by 16S rRNA gene sequencing. We extracted bacterial DNA, amplified it with universal primers targeting variable region 4 of the 16S rRNA gene, sequenced amplicons by Illumina sequencing by synthesis technology using MiSeq, and classified the sequences to bacterial genus. To isolate live bacteria, we used our novel Expanded Quantitative Urine Cultivation protocol (EQUC).[2]
Two cohorts of women with no evidence of clinical UTI provided catheterized urine: 1) women with UUI before UUI treatment and 2) asymptomatic controls undergoing benign gynecologic surgery. Symptoms were assessed by OAB and PFDI questionnaires. To characterize the microbiome, we screened urine for bacterial DNA by 16S rRNA gene sequencing. We extracted bacterial DNA, amplified it with universal primers targeting variable region 4 of the 16S rRNA gene, sequenced amplicons by Illumina sequencing by synthesis technology using MiSeq, and classified the sequences to bacterial genus. To isolate live bacteria, we used our novel Expanded Quantitative Urine Cultivation protocol (EQUC).[2]
Results
Bacterial DNA was amplified and subsequently sequenced from 64% (23/36) of UUI women and 66% (25/38) of controls. The microbiome tended to be dominated by one genus (> 50% total sequence reads). At the genus level, the sequence profiles (urotypes) of both cohorts were similar but not identical. Lactobacillus and Gardnerella urotypes were common in both, but the ratios differed, and other urotypes were seen in only one cohort. These sequences reflect live bacteria, as EQUC identified bacteria from the genera detected by sequencing.
The difference in sequence profile was verified by EQUC. A significantly greater number of bacteria were cultured from women with UUI (p < 0.0001). This difference was due primarily to the genera Actinobaculum, Actinomyces, Aerococcus, Arthrobacter, Corynebacterium, Gardnerella, Oligella, Staphylococcus, and Streptococcus, which were cultured more frequently from women with UUI than from controls. Some bacteria were isolated only from UUI patients.
UUI participants differed from controls in mean age (50 vs. 63, p < 0.05), post-menopausal status (p < 0.05), and coronary artery disease (p=0.02).
Interpretation of Results
The number and type of urinary bacteria differ between women with UUI versus controls and some bacteria were strongly associated with UUI. Information from both DNA sequencing and EQUC contribute to a broader understanding of the urinary microbiome. While UUI status is a likely differentiating factor, the described bacterial differences could also be attributed to other variables, such as age.
Concluding Message
The number and type of urinary bacteria differ between women with UUI versus controls and some bacteria were strongly associated with UUI.
References:
- Wolfe et al. 2012. Evidence of uncultivated bacteria in the adult female bladder. J Clin Microbiol. 50(4):1376-83.
- Hilt et al. 2014. Urine is not sterile: use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. J Clin Microbiol. 52(3):871-6
Disclosures
Funding: This study was supported in part by Astellas Medical and Scientific Affairs and is registered at www.clinicaltrials.gov as NCT01642277. Clinical Trial: Yes Registration Number: This study was supported in part by Astellas Medical and Scientific Affairs and is registered at www.clinicaltrials.gov as NCT01642277. RCT: No Subjects: HUMAN Ethics Committee: IRB Helsinki: Yes Informed Consent: Yes
1Loyola University Chicago


