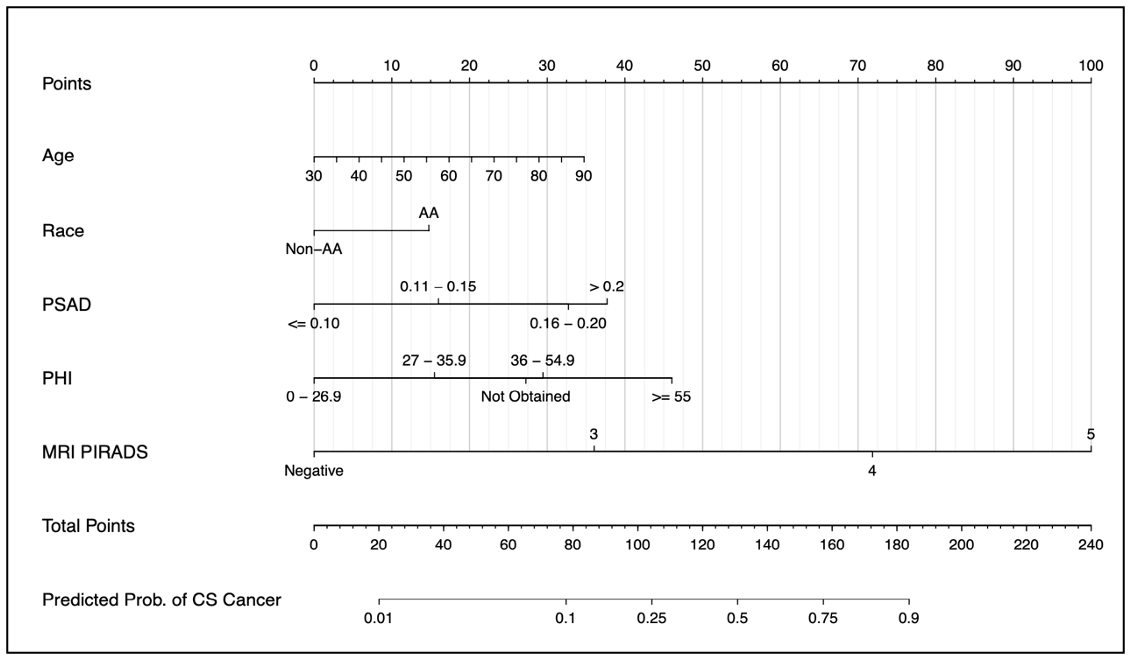
Figure 1: my nMRIsk Prostate Cancer Nomogram using PHI and MRI for prediction of ≥GG2 PCa
Historic risk calculators only include clinical variables such as age, PSA, and DRE, and more recent risk calculators (PCRC-MRI, PLUM) have incorporated mpMRI information. However, there have not been any risk calculators developed that incorporate both advanced serum biomarker and mpMRI. Our hypothesis is that by combining advanced screening methods, we could create risk calculators that have improved csPCa discrimination. Furthermore, while other risk calculators include a significant proportion of prior negative patients at inherently lower risk for csPCa, we focused on a biopsy naïve population.
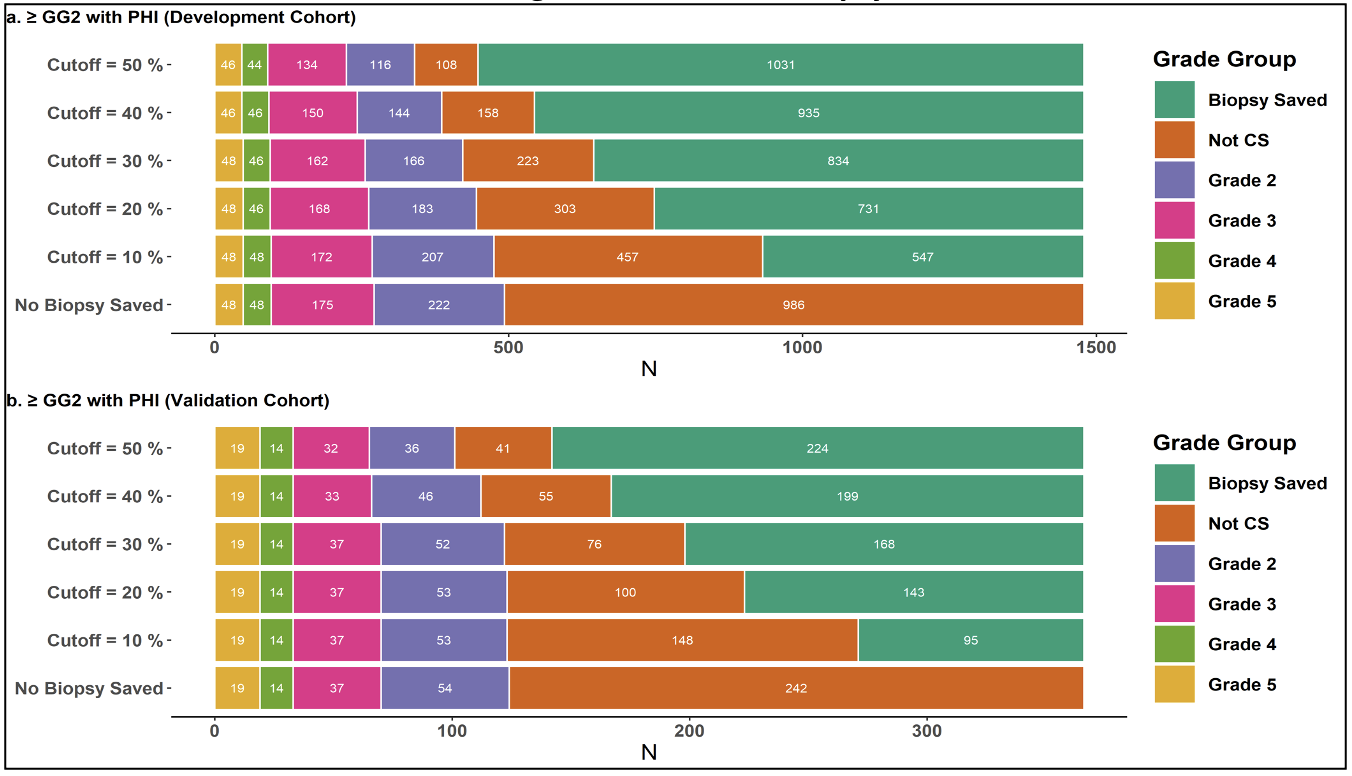
Figure 2: Biopsies Saved for PHI with GG2 in development and validation cohorts. The ‘cut-off’ denotes the risk of finding ≥GG2 disease on biopsy
We recognize that while MRI may be widely used, PHI is only used at select institutions so we generated a series of 6 nomograms, depending on serum biomarker testing available (PHI, % free PSA, or total PSA) and looking at csPCa and ≥GG3 PCa. We identified 1494 biopsy-naïve men who presented to the Northwestern Health system for elevated PSA (2-20ng/mL) evaluation between May 2018-June 2021 and had a separate independent validation cohort of 366 patients who presented to our healthcare system between July 2021-Feb 2022. In our training cohort, 1031 (69%) underwent biopsy with 493 (47.8%) found to have csPCa. The factors predictive of having csPCa on multivariable regression analysis were age, Black race, PHI, PSA density (PSAD), and the highest PIRADS score. The nomograms were highly accurate with Areas Under the Curve (AUC) ranging from 0.885-0.923 between training and validation cohorts. The biopsy outcomes were reported by the probability threshold of having csPCa on biopsy (10%, 20%, 30%, 40%, and 50%). If we assume a 20% biopsy threshold using the csPCa model with PHI in the training cohort, the risk calculator reduces the number of biopsies by 49% while missing 9.7% of csPCa. Looking at the independent validation cohort with a 20% biopsy threshold in the PHI model for prediction of csPCa, the nomogram outperformed our expectations by reducing the number of biopsies by 39.1% (143/366) while only missing 0.8% (1/124) of csPCa.
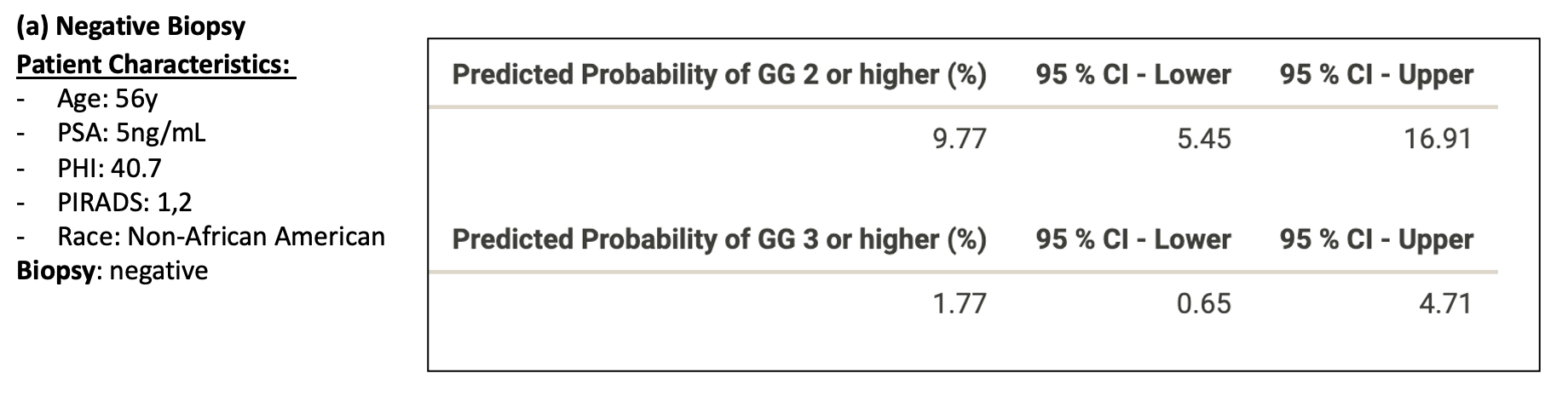
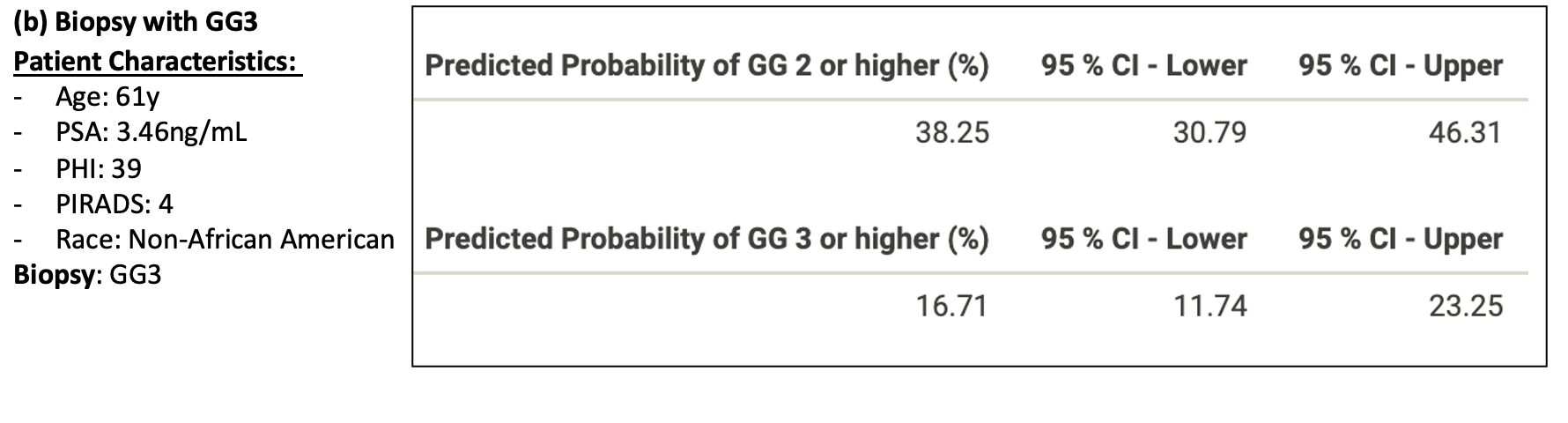
Figure 3: Patient examples of Nomogram Performance with the following biopsy outcomes (a) Negative, (b) Grade Group 3 (GG3)
Written by: Mohammad R Siddiqui, MD, Eric V Li, MD, & Ashley Ross, MD, PhD
Department of Urology, Feinberg School of Medicine, Northwestern University, Chicago, IL
Read the Abstract


