During the last decade, the interdisciplinary meetings of the STITCH project have allowed physicians to understand the wide possibilities offered in the field of research by cooperation with data scientists and the informatics/engineering world. Our project on prostate cancer diagnostics was born from one of these meetings, in which the complex and heterogeneous etiopathology and disease course of this tumor was brainstormed among different health professionals. This led to the first perspective article from the group on the emerging awareness of the benefits that prostate cancer screening research would have gained by applying network medicine, which today translated into a “real-world” prospective applications of network analysis approaches in order to identify meaningful biomarkers for prostate cancer early detection.1
Why we started: The research started surely because of the Global Cancer Observatory data, which illustrates how both the incidence and mortality of prostate cancer (PC) are expected to increase in the next 20 years.2 Based on this premise, recommendations on the diagnostic pathway for prostate cancer have been proposed to the European Commission by members of the European Society of Urology. The proposal today is to attain a risk-adapted strategy not only according to age but also based on biomarkers beyond prostate specific antigen (PSA).3,4 Up to now, many fluid and biopsy-based genomic tests are available for prostate cancer patients in different settings, for risk of disease and treatment outcomes; however today, one biomarker cannot be recommended over another, none is recommended to be used as first line, and they should be considered as one piece of the puzzle in counseling patients.5
That is why we decided to base our work on the identification of meaningful biomarkers in network medicine, since it is a discipline, based on the network science, able to identify the causes of diseases and it defines aggregations clusters with similar characteristics by integrating various types of data, ranging from molecular to clinical and imaging data.
What we did and what we have found: After a pilot cohort study performed in 2021 in which we found a specific cluster of microRNAs correlated to imaging biomarkers and differentially expressed in patients with clinically significant PC, we decided to re-evaluate the microRNAs profiled in our previous study by means of network analysis.6 Specifically, an additional data cleaning process was conducted to remove from each data matrix those microRNAs with a mean value smaller than 50 in each analyzed patients’ group and an interquartile range value smaller than the 15th percentile of the IQR frequency distribution. From this, we found that miR-302a-5p and miR-367-3p were differentially expressed circulating biomarkers, and we further validated it on a cohort of 163 patients. Decision Curve Analysis (DCA) was exploited to assess the utility of different models for decision making by incorporating clinical consequences, such as the benefit of finding disease early or the harm of unnecessary further testing, across a reasonable range of risk thresholds.7 We set as outcome measurement the need for biopsy for csPCa and tested different diagnostic pathway models. The diagnostic pathways performance was evaluated by comparing the number of unnecessary biopsies avoided with a correct diagnosis, under-diagnosis, and over-diagnosis. The proposed pathways are shown in the table below:
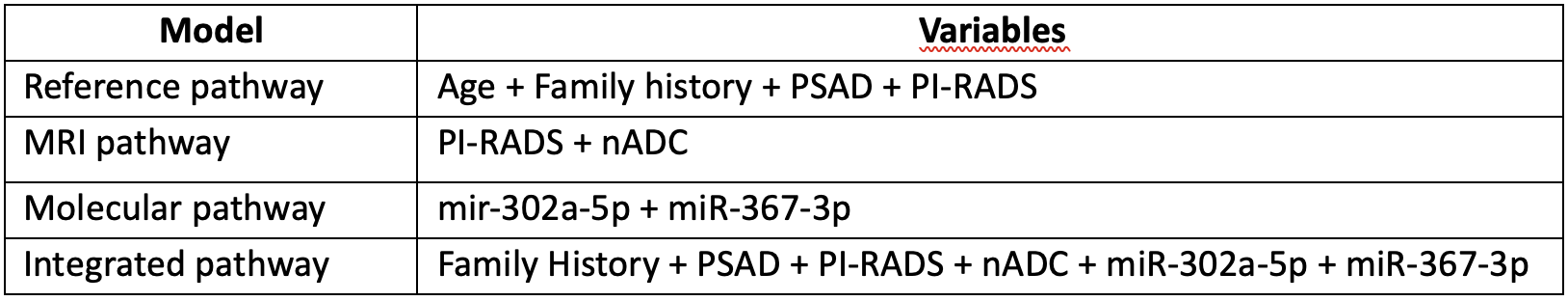
PSAD, PSA density; nADC, normalized Apparent Diffusion Coefficient
What we finally found was that the Integrated pathway allowed for an accurate patient allocation to biopsy, reducing overdiagnosis and overtreatment of clinically insignificant PC.8Where are we going: We validated the predictive role of the proposed “Integrated pathway” by improving the clinical and MRI biomarkers with the addition of circulating molecular markers belonging to the miR-302/miR-367 cluster, whose members' up-regulation has been described in PC tissues and cells.9
Particular attention should be given to how we found without hypnotizing it, that miR-367-3p resulted to be useful in discriminating between Gleason Grade (GG)4 and GG5 with respect to other categories, providing for the first time a way of classifying pre-operatively cancer aggressiveness. Also, we observed how miR-302a-5p was differentially expressed in GG2 tumors, and we believe that it could be used to discriminate between GG2 and other categories, providing for the first time a way of classifying pre-operatively cancer aggressiveness. It has already been shown in literature how miR-302a albeit pivotal for csPCa cell growth, is inversely expressed in higher Gleason score PCa.10 In the near future we aim at confirming it on a larger sample size.
Written by: Martina Pecoraro,1 Giuseppina Catanzaro,2 Federica Conte,3 Zein Mersini Besharat,2 Emanuele Messina,1 Ludovica Laschena,1 Sofia Trocchianesi,4 Elena Splendiani,4 Alessandro Sciarra,5 Carlo Catalano,1 Paola Paci,6 Elisabetta Ferretti,2 Valeria Panebianco7
- Department of Radiological Sciences, Oncology and Pathology, Sapienza University, Policlinico Umberto I, Rome, Italy.
- Department of Experimental Medicine, Sapienza University, Policlinico Umberto I, Rome, Italy.
- Institute for Systems Analysis and Computer Science "A. Ruberti" (IASI), National Research Council (CNR), Rome, Italy.
- Department of Molecular Medicine, Sapienza University of Rome, Rome, Italy.
- Department of Maternal Infant and Urologic Sciences, Sapienza University of Rome, Rome, Italy.
- Department of Computer, Control and Management Engineering, Sapienza University, Rome, Italy.
- Department of Radiological Sciences, Oncology and Pathology, Sapienza University, Policlinico Umberto I, Rome, Italy.
References:
- Panebianco V, Pecoraro M, Fiscon G, et al (2020) Prostate cancer screening research can benefit from network medicine: an emerging awareness. npj Syst Biol Appl 6:13.
- Sung H, Ferlay J, Siegel RL, et al (2021) Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J Clin 71:209–249.
- Van Poppel H, Hogenhout R, Albers P, et al (2021) Early Detection of Prostate Cancer in 2020 and Beyond: Facts and Recommendations for the European Union and the European Commission. European Urology 79:327–329.
- Van Poppel H, Roobol MJ, Chapple CR, et al (2021) Prostate-specific Antigen Testing as Part of a Risk-Adapted Early Detection Strategy for Prostate Cancer: European Association of Urology Position and Recommendations for 2021. European Urology 80:703–711.
- Cooperberg MR, Carroll PR, Dall’Era MA, et al (2019) The State of the Science on Prostate Cancer Biomarkers: The San Francisco Consensus Statement. European Urology 76:268–272.
- Panebianco V, Paci P, Pecoraro M, et al (2021) Network Analysis Integrating microRNA Expression Profiling with MRI Biomarkers and Clinical Data for Prostate Cancer Early Detection: A Proof of Concept Study. Biomedicines 9:1470.
- Vickers AJ, Elkin EB (2006) Decision Curve Analysis: A Novel Method for Evaluating Prediction Models. Med Decis Making 26:565–574.
- Pecoraro M, Catanzaro G, Conte F, et al (2023) Prospective Validation Study of a Novel Integrated Pathway Based on Clinical Features, Magnetic Resonance I
- Guo Y, Cui J, Ji Z, et al (2017) miR-302/367/LATS2/YAP pathway is essential for prostate tumor-propagating cells and promotes the development of castration resistance. Oncogene 36:6336–6347.
- Zhang G-M, Bao C-Y, Wan F-N, et al (2015) MicroRNA-302a Suppresses Tumor Cell Proliferation by Inhibiting AKT in Prostate Cancer. PLoS ONE 10:e0124410.


