In metastatic prostate cancer, the proportion of tumor-derived cfDNA in blood (i.e. circulating tumor DNA (ctDNA) fraction) strongly correlates with outcomes in multiple clinical contexts. This includes the depth of ctDNA fraction decline after systemic treatment initiation (relative to baseline levels),4,5 as well as the amount of ctDNA measured at clinical progression prior to initiating next line of therapy.6–9 However, the prognostic potential of baseline pretreatment ctDNA fraction and its relationship to established clinical prognostic factors had not been previously tested in a large contemporary clinical cohort with standardized outcomes assessment.
We measured plasma ctDNA fraction in 491 patients with mCRPC (plus serial samples) with comprehensive clinical annotation. Key findings:
1. ctDNA fraction was strongly associated with initial PSA response to systemic therapy, time to PSA progression, and overall survival—including after adjustment for recognized clinical prognostic covariables and independent of first- or second-line context. For example, high ctDNA (>30%) at initiation of first-line mCRPC therapy was linked to a 5.6 times greater risk of death in univariable analysis (versus patients with <2% ctDNA; p<0.001). These results reaffirm that ctDNA fraction is a powerful independent predictor of outcomes and suggest a potential role for baseline ctDNA fraction as a stratification factor in future clinical trials.
2. ctDNA fraction also largely determines whether widely available liquid biopsy biomarker tests (e.g. for BRCA2 mutations) will be informative.10 Cancer genomic alterations cannot be sensitively detected if ctDNA fraction is low (i.e. resulting in false negative test results), and previously there were no practical tools to determine prior to blood draw whether a patient has sufficient ctDNA fraction for informative ctDNA genotyping. We built a freely available online tool (ctdna.org) to predict whether patients with progressive mCRPC have sufficient ctDNA fraction for ctDNA genomic testing, leveraging routinely surveyed clinical characteristics that correlate with ctDNA levels. We anticipate that ctdna.org will help users decide on whether to pursue ctDNA or alternative biomarker tests (e.g. archival tissue, fresh metastatic biopsy, or germline-only screening), based on predicted patient ctDNA fraction.
ctDNA profiling is minimally invasive, relatively inexpensive, and already incorporated into the standard of care for patients with metastatic prostate cancer. Our study helps advocate that ctDNA fraction can be used for patient risk stratification and additionally offers a practical framework (Figure 1) for expedient informative biomarker testing.11
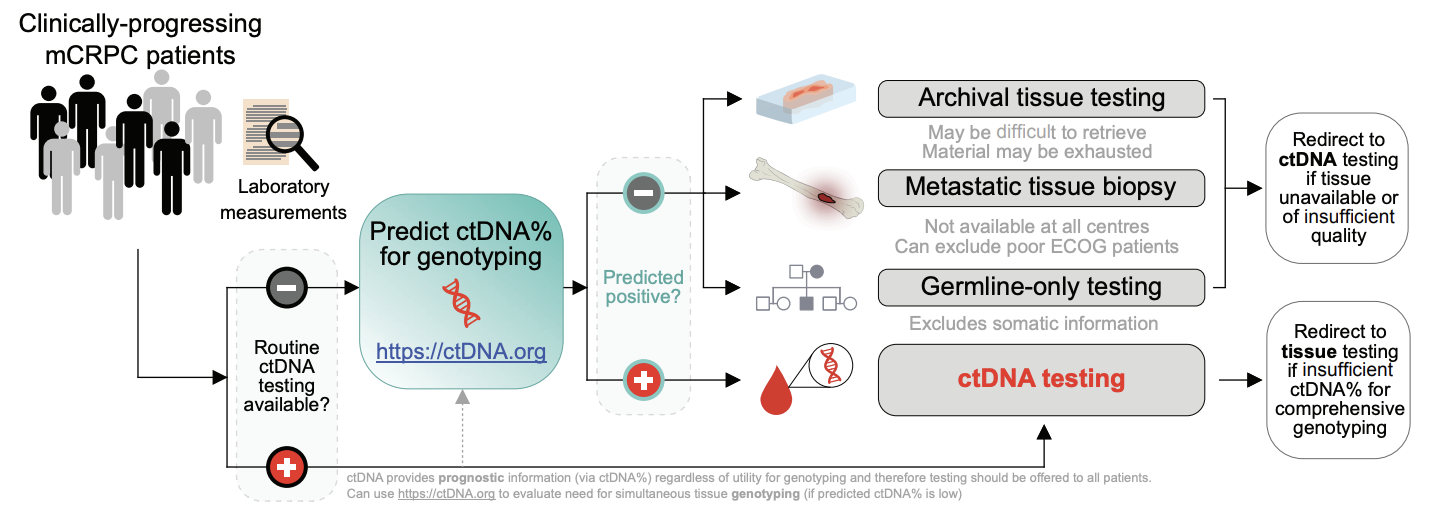
Figure 1. Proposed workflow for optimized clinical biomarker profiling for mCRPC incorporating ctDNA testing.
The ctDNA prediction tool (available at http://ctDNA.org) can be used to inform optimal strategy for mCRPC genomic biomarker testing, offering guidance as to whether to pursue ctDNA or tissue-based genotyping (in resource-limited circumstances where both testing modalities cannot be pursued simultaneously). Tissue-based genotyping should be initiated for patients with low predicted ctDNA fraction. However, ctDNA testing also offers valuable prognostic information (via ctDNA fraction) regardless of ctDNA fraction sufficiency for sensitive genotyping and therefore should be offered to patients if available as a prognostic adjunct (potentially in tandem with tissue-based genotyping). The tool’s output includes the probability of a sample having ctDNA≥2% and a point estimate of the predicted plasma ctDNA fraction. Finally, the ctDNA fraction prediction tool is flexible to any combination of missing data as well as differences in laboratory reference range values for lactate dehydrogenase and alkaline phosphatase. Figure and caption modified from Fonseca et al., Nature Communications (2024) under a Creative Commons Attribution 4.0 International License.
Written by:
- Cameron Herberts, PhD, Vancouver Prostate Centre, Department of Urologic Sciences, University of British Columbia, Vancouver, BC, Canada.
- Nicolette M. Fonseca, PhD, Vancouver Prostate Centre, Department of Urologic Sciences, University of British Columbia, Vancouver, BC, Canada.
- Corinne Maurice-Dror, MD, Department of Medical Oncology, BC Cancer, Vancouver, BC, Canada
- Matti Annala, MSc, Vancouver Prostate Centre, Department of Urologic Sciences, University of British Columbia, Vancouver, BC, Canada, Prostate Cancer Research Center, Faculty of Medicine and Health Technology, Tampere University and Tays Cancer Center, Tampere, Finland.
- Kim N. Chi, MD, Vancouver Prostate Centre, Department of Urologic Sciences, University of British Columbia, Vancouver, BC, Canada, Department of Medical Oncology, BC Cancer, Vancouver, BC, Canada
- Alexander W. Wyatt, DPhil, Vancouver Prostate Centre, Department of Urologic Sciences, University of British Columbia, Vancouver, BC, Canada, Michael Smith Genome Sciences Centre, BC Cancer, Vancouver, BC, Canada
- Swami, U., McFarland, T. R., Nussenzveig, R. & Agarwal, N. Advanced Prostate Cancer: Treatment Advances and Future Directions. Trends Cancer Res. 6, 702–715 (2020).
- Halabi, S. et al. Updated prognostic model for predicting overall survival in first-line chemotherapy for patients with metastatic castration-resistant prostate cancer. J. Clin. Oncol. 32, 671–677 (2014).
- Chi, K. N. et al. A prognostic index model for predicting overall survival in patients with metastatic castration-resistant prostate cancer treated with abiraterone acetate after docetaxel. Ann. Oncol. 27, 454–460 (2016).
- Tolmeijer, S. H. et al. Early On-treatment Changes in Circulating Tumor DNA Fraction and Response to Enzalutamide or Abiraterone in Metastatic Castration-Resistant Prostate Cancer. Clin. Cancer Res. 29, 2835–2844 (2023).
- Jayaram, A. et al. Plasma tumor gene conversions after one cycle abiraterone acetate for metastatic castration-resistant prostate cancer: a biomarker analysis of a multicenter international trial. Ann. Oncol. 32, 726–735 (2021).
- Nørgaard, M. et al. Prognostic Value of Low-Pass Whole Genome Sequencing of Circulating Tumor DNA in Metastatic Castration-Resistant Prostate Cancer. Clin. Chem. 69, 386–398 (2023).
- Annala, M. et al. Circulating Tumor DNA Genomics Correlate with Resistance to Abiraterone and Enzalutamide in Prostate Cancer. Cancer Discov. 8, 444–457 (2018).
- Kohli, M. et al. Clinical and genomic insights into circulating tumor DNA-based alterations across the spectrum of metastatic hormone-sensitive and castrate-resistant prostate cancer. EBioMedicine 54, 102728 (2020).
- Sumanasuriya, S. et al. Elucidating Prostate Cancer Behaviour During Treatment via Low-pass Whole-genome Sequencing of Circulating Tumour DNA. Eur. Urol. 80, 243–253 (2021).
- Herberts, C. & Wyatt, A. W. Technical and biological constraints on ctDNA-based genotyping. Trends Cancer Res. 7, 995–1009 (2021).
- Fonseca, N. M. et al. Prediction of plasma ctDNA fraction and prognostic implications of liquid biopsy in advanced prostate cancer. Nat. Commun. 15, 1828 (2024).


