Polygenic risk scores (PRS) have been developed by many groups that combine these variants into a single score representing the likelihood of an individual being diagnosed with prostate cancer. However, few studies have externally validated published PRS and directly compared their performance. In addition, little is known whether PRS can be used as a marker of clinically significant prostate cancer.
Our study sought to evaluate the performance of published PRS constructs on an external dataset, both for prostate cancer incidence and aggressiveness. We used PRS constructs published in the Polygenic Score Catalog to perform this analysis.1 These constructs were used to derive PRS on patients within the Michigan Genomics Initiative (MGI), a biorepository prospectively maintained and linked to the patient’s electronic medical record.2 To evaluate the association of PRS and prostate cancer incidence, we focus on unrelated male adults of inferred European ancestry, which identified 15,310 controls and 2,740 prostate cancer cases.
We assessed the performance of PRS on predicting prostate cancer aggressiveness; all previously identified cases were cross-referenced within the Michigan Urologic Surgery Improvement Collaborative (MUSIC), a statewide collaborative across all practice types that prospectively collects demographic and clinical variables.3 Based on the National Comprehensive Cancer Network risk stratification cases were categorized into low, intermediate, or high risk. We identified 1,174 participants with a prostate biopsy within MUSIC and MGI, with 293 low-risk, 638 intermediate-risk, and 243 high-risk individuals. A sensitivity analysis was performed using 1,007 participants with prostatectomy pathology data from MUSIC (58 low-risk, 707 intermediate-risk, and 242 high-risk).
There was a strong correlation with prostate cancer incidence across these PRS, with adjusted AUC between 0.519 and 0.669. There was also a strong correlation between higher PRS and increasing prostate cancer incidence (Fig 1).
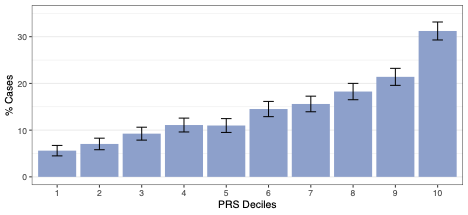
Figure 1. Overall prostate cancer case distribution across PRS risk deciles.
The top three PRS were then evaluated for the ability to distinguish between disease aggressiveness by dividing participants into deciles based on PRS. There was no association between PRS and disease aggressiveness using prostate biopsy-based risk categories, as noted by the nearly identical PRS distribution across the risk categories for all three PRS (Fig 2).

Figure 2. Density curves for top-performing polygenic risk scores PRS11, PRS12, and PRS15 across biopsy-based prostate cancer grades and control subjects. Corresponding deciles within the subsets are color-coded and indicated by vertical lines.
A validation for the top-performing PRS using prostatectomy pathology-based risk categories did not show any difference between PRS distribution across the NCCN risk categories (Fig 3).

Figure 3. Density curves of PCaPRS11 across pathology-based prostate cancer risk groups versus controls. Corresponding deciles within the subsets are color-coded and designated by vertical lines.
Our findings regarding the prediction of any disease development align with the original studies for PRS, with higher scores correlating to a higher likelihood of prostate cancer incidence. However, at present, there is limited predictive ability to discern aggressive disease, and further work is ongoing to incorporate prostate cancer risk groups into PRS development.4 Klein et al. similarly did not find an association between PRS and aggressive prostate cancer in men of European descent; in fact, PSA performed better as a predictor of prostate cancer metastasis or mortality compared to PRS (AUC 0.78 vs. 0.63).5 It appears that the solution also does not lie in simply increasing the number of SNPs captured by constructing a genome-wide PRS, as recently reported by Darst et al.6 Newer PRS aim to evaluate more diverse populations and high-risk disease; for example, Chen et al. developed a PRS in men of African ancestry and found an OR of 1.23 for aggressive prostate cancer relative to those in the 40-60% PRS category in a case-case analysis.7 The most promising data appears to be the use of PRS in combination with other clinical variables such as family history, age, and PSA values, which may improve the detection of clinically significant prostate cancer and reduce the number of screening biopsies.4,8
Written by: Helen Sun, MD, & Randy Vince Jr., MD, MS, Urology Institute, University Hospitals of Cleveland, Case Western Reserve University, Cleveland, OH
References:
- Lambert SA, Gil L, Jupp S, et al. The Polygenic Score Catalog as an open database for reproducibility and systematic evaluation. Nat Genet. 2021;53(4):420-425. doi:10.1038/s41588-021-00783-5
- Fritsche LG, Gruber SB, Wu Z, et al. Association of Polygenic Risk Scores for Multiple Cancers in a Phenome-wide Study: Results from The Michigan Genomics Initiative. The American Journal of Human Genetics. 2018;102(6):1048-1061. doi:10.1016/j.ajhg.2018.04.001
- Riedinger CB, Womble PR, Linsell SM, et al. Variation in Prostate Cancer Detection Rates in a Statewide Quality Improvement Collaborative. Journal of Urology. 2014;192(2):373-378. doi:10.1016/j.juro.2014.02.088
- Aly M, Wiklund F, Xu J, et al. Polygenic Risk Score Improves Prostate Cancer Risk Prediction: Results from the Stockholm-1 Cohort Study. European Urology. 2011;60(1):21-28. doi:10.1016/j.eururo.2011.01.017
- Klein RJ, Vertosick E, Sjoberg D, et al. Prostate cancer polygenic risk score and prediction of lethal prostate cancer. npj Precis Onc. 2022;6(1):1-8. doi:10.1038/s41698-022-00266-8
- Darst BF, Shen J, Madduri RK, et al. Evaluating approaches for constructing polygenic risk scores for prostate cancer in men of African and European ancestry. Am J Hum Genet. 2023;110(7):1200-1206. doi:10.1016/j.ajhg.2023.05.010
- Chen F, Madduri RK, Rodriguez AA, et al. Evidence of Novel Susceptibility Variants for Prostate Cancer and a Multiancestry Polygenic Risk Score Associated with Aggressive Disease in Men of African Ancestry. European Urology. Published online March 2023:S0302283823025617. doi:10.1016/j.eururo.2023.01.022
- Huynh-Le MP, Karunamuni R, Fan CC, et al. Prostate cancer risk stratification improvement across multiple ancestries with new polygenic hazard score. Prostate Cancer Prostatic Dis. 2022;25(4):755-761. doi:10.1038/s41391-022-00497-7


