Yet, how is this mediated? One hypothesis is that the gut microbiome may mediate the effects of TRF.
• Serum metabolomics suggested indirectly that the gut microbiome was different
• Gut microbiome contributes to obesity and dysmetabolism
• However, until recently, the gut microbiome was thought to be stable over long periods of time
• More recent evidence suggests diet changes the gut microbiome rapidly
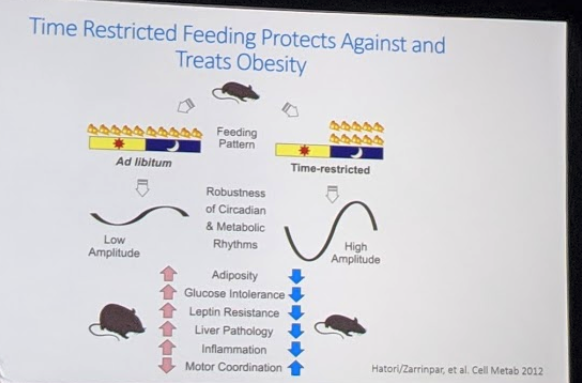
In their more recent work, his group conducted a mouse study that compared 3 groups of mice: a wild type mouse type that was on a “normal” eating schedule, an ad-libitum (access to food all the time) diet of choice population, and an TRF population.
• In the normal control population, they found that food intake peaked overnight (as expected). They found that 15% of the gut microbiome species (representing 42% of the population) cycled during the 24-hour period. There were distinct cell populations that peaked at different parts of the 24-hour cycle, relative to the fasting and eating periods:
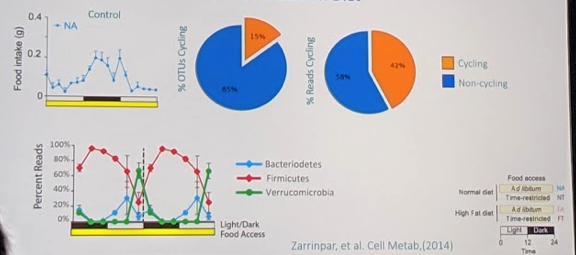
In the diet of choice group, there was food intake throughout the day. However, there was much less cycling of microbiome species (7% of species representing 20% of the population). And no real distinct peaks (see bottom left):
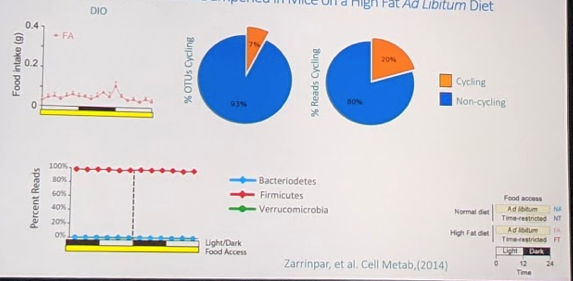
In the TRF population, there was a peak in food intake at the time of timed eating. However, surprisingly, there was much less cycling of microbiome species (7% of species representing 12% of the population). And no real distinct peaks (see bottom left):
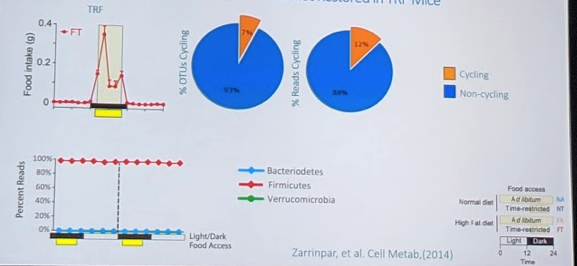
The TRF and ad-libitum diets were compositionally similar. On further analysis, however, they noted that TRF mice excreted more bile acids that the ad-libitum mice.
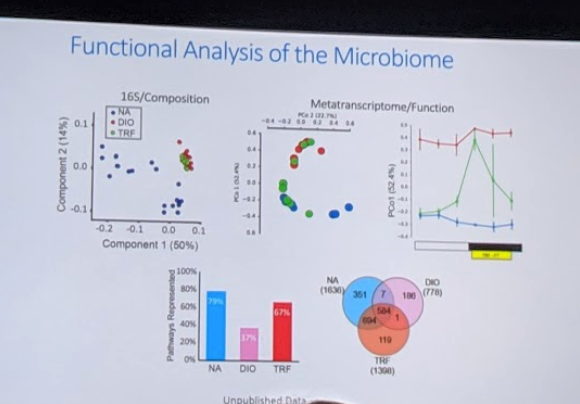
In order to sort out why there wasn’t more cycling in the TRF population, they decided to look at RNA expression in the mice rather than DNA expression – so that they could get a better sense of the functional expression of genes. Interestingly, in this analysis they found the truth was that TRF mice had gene expression that overlapped with normal mice and ad-libitum mice – they were a hybrid!
Based on this, his last section was their most recent work to engineer a bacteria that could colonize conventionally raised wild type mice. They were able to do successfully with durable colonization. More importantly, they were able to impart important physiologic changes with the addition of this one new cell line! This has huge implications for management of many chronic conditions and cancer!
Speaker: Amir Zarrinpar, MD, PhD
References:
All slides and images are from Amir Zarrinpar, MD, PhD
Written by: Thenappan Chandrasekar, MD, Clinical Instructor, Thomas Jefferson University, @tchandra_uromd, @TjuUrology, at the 19th Annual Meeting of the Society of Urologic Oncology (SUO), November 28-30, 2018 – Phoenix, Arizona


