Prostate cancer is often detected initially by a rise in serum prostate specific antigen (PSA) and since PSA was recommended for screening by the FDA in 1994 the incidence of prostate cancer has increased dramatically (2).. Although the front line treatments for prostate cancer (which include radical prostatectomy, radiation and androgen deprivation therapy) are initially effective, they are also associated with significant side effects. These side effects include urinary incontinence, erectile dysfunction and bowel dysfunction and they can impact negatively on the patient’s quality of life.
Furthermore, it is widely acknowledged that most men die with prostate cancer rather than because of the disease and large scale studies have failed to provide definitive evidence for the overall benefits of PSA screening (3). For illustration, of 1000 men screened, the PSA test will produce a false positive in 100-120 cases. Of the approximately 110 men found to have elevated PSA and a subsequently confirmed diagnosis of prostate cancer, more than half will face significant side effects of treatment and just 4-5 men will die from prostate cancer (4). In its most favourable interpretation this data reveals that screening the 1000 men will have saved about one life while impacting negatively on the quality of life of well over 100 (http://prostatecanceruk.org).
So a key conclusion from these and many other observations is that, while the minority of patients who have life-threatening disease do benefit from the important developments in novel therapies and 'management' strategies that have been made recently, many patients with clinically insignificant disease suffer from over-treatment. In part, it was in for these reasons that in 2012 the USPTF recommended against PSA screening. This recommendation resulted in a 28% decrease in the incidence of prostate cancer in the following year. However, this recommendation is also leading to a change in the stage at which men initially present. For example, although the figures have been disputed, it has been reported that the number of cases of metastatic prostate cancer rose by 72% between 2004 and 2013 as consequence of the USPTF ruling.
So, for a disease that in most cases is not threatening to a man’s natural lifespan, the decision on how best to manage it should be made with careful consideration of the lasting impact of treatment on the patient's daily life. It is now increasingly clear that active surveillance is a viable option for many men who are diagnosed with prostate cancer (5–7). This non-invasive alternative to treatment is recommended for men with organ confined non-aggressive prostate cancer although international recommendations (AUA, EAU) vary in both eligibility criteria and follow up modalities (6). Although many studies have reported that active surveillance is as effective as conventional treatment for the management of indolent prostate cancer, many physicians and patients are reluctant to opt for this strategy (5). It has been suggested that this is, in part, due to a lack of suitable tools for the diagnosis and routine monitoring of the disease - tools that give a reliable indication of the likely clinical outcome (6).
Despite its perceived utility as a biomarker for prostate cancer, PSA measurements fail to provide sufficient evidence of the severity or potential aggressiveness of prostate cancer and it is therefore not well suited to supporting the early treatment decisions outlined above. There are a number of other clinical ‘tools’ available to help stratify patients based on their presumed degree of risk for developing aggressive Prostate cancer – most notably the Gleason scoring system (8). This system relies on the pathological scoring of patient biopsy samples and is limited to some extent by known sampling errors and challenges in the assessment of tissue pathology. Effective management of Prostate cancer patient treatment decisions would benefit greatly from the availability of specific molecular biomarkers. A number of novel tissue gene-based tests have been developed and are available. However, these suffer from the same limitations as the Gleason scoring system as they also rely on access to and interpretation of tissue samples.
So, for a disease that in most cases is not threatening to a man’s natural lifespan, the decision on how best to manage it should be made with careful consideration of the lasting impact of treatment on the patient's daily life. It is now increasingly clear that active surveillance is a viable option for many men who are diagnosed with prostate cancer (5–7). This non-invasive alternative to treatment is recommended for men with organ confined non-aggressive prostate cancer although international recommendations (AUA, EAU) vary in both eligibility criteria and follow up modalities (6). Although many studies have reported that active surveillance is as effective as conventional treatment for the management of indolent prostate cancer, many physicians and patients are reluctant to opt for this strategy (5). It has been suggested that this is, in part, due to a lack of suitable tools for the diagnosis and routine monitoring of the disease - tools that give a reliable indication of the likely clinical outcome (6).
Despite its perceived utility as a biomarker for prostate cancer, PSA measurements fail to provide sufficient evidence of the severity or potential aggressiveness of prostate cancer and it is therefore not well suited to supporting the early treatment decisions outlined above. There are a number of other clinical ‘tools’ available to help stratify patients based on their presumed degree of risk for developing aggressive Prostate cancer – most notably the Gleason scoring system (8). This system relies on the pathological scoring of patient biopsy samples and is limited to some extent by known sampling errors and challenges in the assessment of tissue pathology. Effective management of Prostate cancer patient treatment decisions would benefit greatly from the availability of specific molecular biomarkers. A number of novel tissue gene-based tests have been developed and are available. However, these suffer from the same limitations as the Gleason scoring system as they also rely on access to and interpretation of tissue samples.
Consequently, there is a desire to develop tests that can be performed using more accessible biofluids such as urine and blood. Although a number of urine-based gene/protein measurements have been implemented as clinical diagnostic tools (9–11), novel serum-based biomarkers remain confined to the kallikrein family of proteins – as in tests like the 4Kscore which is a combined measurement of total PSA, free PSA, intact PSA and human kallikrein-related peptide 2 (hK2) (12). Although the serum and urine tests described in this review are certainly promising a significant challenge to overcome is that of measuring early stage tumour genes /proteins when they are diluted extensively in fluids such as serum and urine. For this reason, the choice of technology used to discover and measure markers of interest will be a key consideration for the successful development and clinical implementation of fluid-based prostate cancer biomarkers.
For the compelling reasons outlined above, there has been significant recent interest in the development of novel protein biomarkers. The process of discovering and developing new protein biomarkers is often conducted in stages (13) and requires technologies that will allow for fast and consistent identification of proteins spanning the expansive dynamic range of the proteome. The selection of an appropriate sample material for which to base initial discovery experiments are influenced by considerations that are outlined in this review.
Proteome scale technologies have improved significantly in recent years and despite limitations, mass spectrometry has emerged as a key enabling technology for biomarker discovery such that it is now possible to use it to simultaneously identify and quantify thousands of proteins in biological samples. Strategies for evaluating the performance of candidate biomarkers, which previously relied on antibody-based approaches such as ELISA, now include a range of nano- and aptamer-based technologies that make it possible to undertake multiplexed measurement of low abundance proteins in biological fluids. At the same time a range of 'targeted' mass spectrometry methods can now be used for candidate biomarker evaluation.
For the compelling reasons outlined above, there has been significant recent interest in the development of novel protein biomarkers. The process of discovering and developing new protein biomarkers is often conducted in stages (13) and requires technologies that will allow for fast and consistent identification of proteins spanning the expansive dynamic range of the proteome. The selection of an appropriate sample material for which to base initial discovery experiments are influenced by considerations that are outlined in this review.
Proteome scale technologies have improved significantly in recent years and despite limitations, mass spectrometry has emerged as a key enabling technology for biomarker discovery such that it is now possible to use it to simultaneously identify and quantify thousands of proteins in biological samples. Strategies for evaluating the performance of candidate biomarkers, which previously relied on antibody-based approaches such as ELISA, now include a range of nano- and aptamer-based technologies that make it possible to undertake multiplexed measurement of low abundance proteins in biological fluids. At the same time a range of 'targeted' mass spectrometry methods can now be used for candidate biomarker evaluation.
One such technique – multiple reaction monitoring (MRM) – provides robust, sensitive, quantitative, specific and high-throughput multiplexed measurement of protein biomarkers in complex bio specimens. Furthermore, MRM is already routinely used in clinical settings and various CLIA-approved MRM-based assays are now available as diagnostic tests (14). So, MRM offers the potential to support the development of new protein biomarker panels (or 'signatures') but it is clear that this will require careful consideration of all aspects of assay development to ensure that such new 'MRM-based' assays meet the requirements of tests that are used for clinical decision-making.
It should go without saying that pre-analytical sample processing, MRM-assay assay development and post-analytical data handling must be undertaken in a manner that meets the requirements for clinical use - potentially across multiple laboratory sites. So, it is evident that new tests are needed in prostatecancer and developing them to the stage that they can be introduced in the clinical setting i.e. are 'fit for purpose', will require the combined and integrated effort of a range of discrete domain experts including those with knowledge and practical experience of hospital laboratory clinical chemistry.
It should go without saying that pre-analytical sample processing, MRM-assay assay development and post-analytical data handling must be undertaken in a manner that meets the requirements for clinical use - potentially across multiple laboratory sites. So, it is evident that new tests are needed in prostatecancer and developing them to the stage that they can be introduced in the clinical setting i.e. are 'fit for purpose', will require the combined and integrated effort of a range of discrete domain experts including those with knowledge and practical experience of hospital laboratory clinical chemistry.
One such effort is underway in the Biomedical Proteomics Research Group in University College Dublin (Ireland; http://www.biomedicalproteomics.org) who are developing MRM assays including for the measurement of a panel of proteins that may be able to discriminate between organ confined and non-organ confined prostate cancer. This test, called the OCProDx test, involves multiplexed measurement of a protein panel in patient serum samples. With input from patients, clinicians, lab scientists and those with clinical diagnostic assay development experience and initial support from the EU’s Horizon2020 programme, the OCProDx test is being developed further by the UCD spin-out Atturos (www.Atturos.com) with the intention of bringing it to clinical use. Further 'fit for purpose' evaluation of this biomarker signature in a dedicated protein biomarker evaluation lab. In a collaborative arrangement with Agilent Technologies this lab is equipped with the latest mass spectrometer for sensitive MRM measurements. This, and the many similar efforts underway elsewhere, may lead to the development and delivery of more effective clinical decision-making tools to support the many men who are diagnosed with prostate cancer.
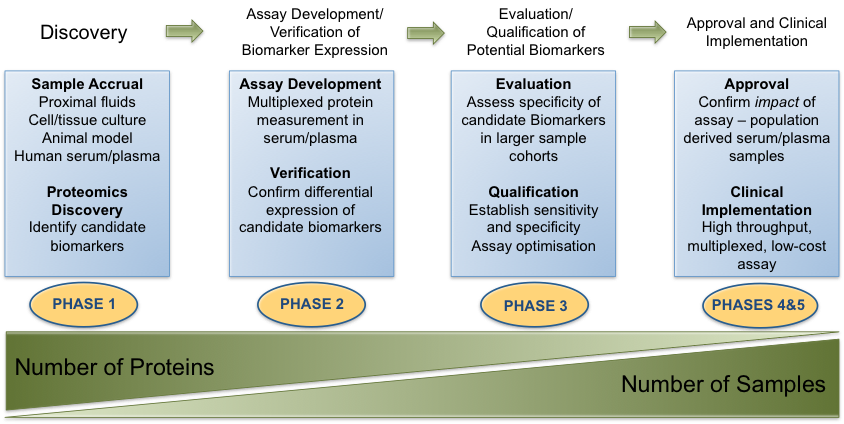
Figure 1: Discovery and Clinical Implementationof Protein Biomarkers: outline of the process involved in bringing candidate protein biomarkers through from the discovery phase, evaluation and ultimately to adoption as a clinical assay for subsequent validation (Figure adapted from Rifai et al, 2006 (13).

Figure 1: Discovery and Clinical Implementationof Protein Biomarkers: outline of the process involved in bringing candidate protein biomarkers through from the discovery phase, evaluation and ultimately to adoption as a clinical assay for subsequent validation (Figure adapted from Rifai et al, 2006 (13).
Written by: Claire L. Tonry, Emma Leacy and Stephen R. Pennington
Read the Abstract
References
1. Nagavarapu U. Prevention and Treatment of Prostate Cancer. 2013:
2. Howrey BT, Kuo Y-F, Lin Y-L, Goodwin JS. The impact of PSA screening on prostate cancer mortality and overdiagnosis of prostate cancer in the United States. [Internet]. J. Gerontol. A. Biol. Sci. Med. Sci. 2013;68(1):56–61.
3. Crawford ED, Bennett CL, Andriole GL, Garnick MB, Petrylak DP. The utility of prostate-specific antigen in the management of advanced prostate cancer. [Internet]. BJU Int. 2013;112(5):548–60.
4. Moyer VA. Screening for Prostate Cancer: U.S. Preventive Services Task Force Recommendation Statement. Ann. Intern. Med. 2012;157(2):120–135.
5. Lund L, Svolgaard N, Poulsen MH. Prostate cancer: a review of active surveillance [Internet]. Res Rep Urol 2014;6:107–112.
6. Bruinsma SM et al. Active surveillance for prostate cancer: a narrative review of clinical guidelines [Internet]. Nat. Rev. Urol. 2016;1–17.
7. Bangma CH et al. Active surveillance for low-risk prostate cancer.. Crit. Rev. Oncol. Hematol. 2013;85(3):295–302.
8. Humphrey PA. Gleason grading and prognostic factors in carcinoma of the prostate [Internet]. Mod. Pathol. 2004;17(3):292–306.
9. Schalken JA, Hessels D, Verhaegh G. New targets for therapy in prostate cancer: Differential display code 3 (DD3PCA3), a highly prostate cancer-specific gene. Urology 2003;62(5 SUPPL. 1):34–43.
10. Cornu JN et al. Urine TMPRSS2: ERG fusion transcript integrated with PCA3 score, genotyping, and Biological features are correlated to the Results of prostatic biopsies in men at risk of prostate cancer. Prostate 2013;73(3):242–249.
11. Van Neste L et al. Detection of High-grade Prostate Cancer Using a Urinary Molecular Biomarker-Based Risk Score. Eur. Urol. 2016;1–9.
12. McDonald ML, Parsons JK. 4-Kallikrein Test and Kallikrein Markers in Prostate Cancer Screening [Internet]. Urol. Clin. North Am. 2016;43(1):39–46.
13. Rifai, Nader Gillette, Michael A Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility [Internet]. Nat. Biotechnol. 2006;24(8):971–983.
14. Baker ES et al. Mass spectrometry for translational proteomics: progress and clinical implications [Internet]. Genome Med. 2012;4(8):63.


