To achieve this, we systematically searched multiple databases, including Cochrane, Ovid, Web of Science, and PubMed, limiting results to articles published between January 1, 2000, and January 30, 2023. Our review included studies that compared gene expression in individuals with urethral stricture to healthy individuals, utilizing advanced molecular techniques to analyze gene expression in blood, urine, or tissue samples.
From our search criteria, we selected four studies for inclusion. These studies utilized complex molecular biology methods to measure and quantify gene expression in various samples. Our comprehensive analysis revealed that the expressions of CXCR3 and NOS2 genes were notably downregulated in urethral tissue samples. Conversely, TGFB1, UPK3A, and CTGF were upregulated in plasma, urine, and urethral tissue samples, respectively, in patients with urethral stricture compared to healthy controls (Figure 1).
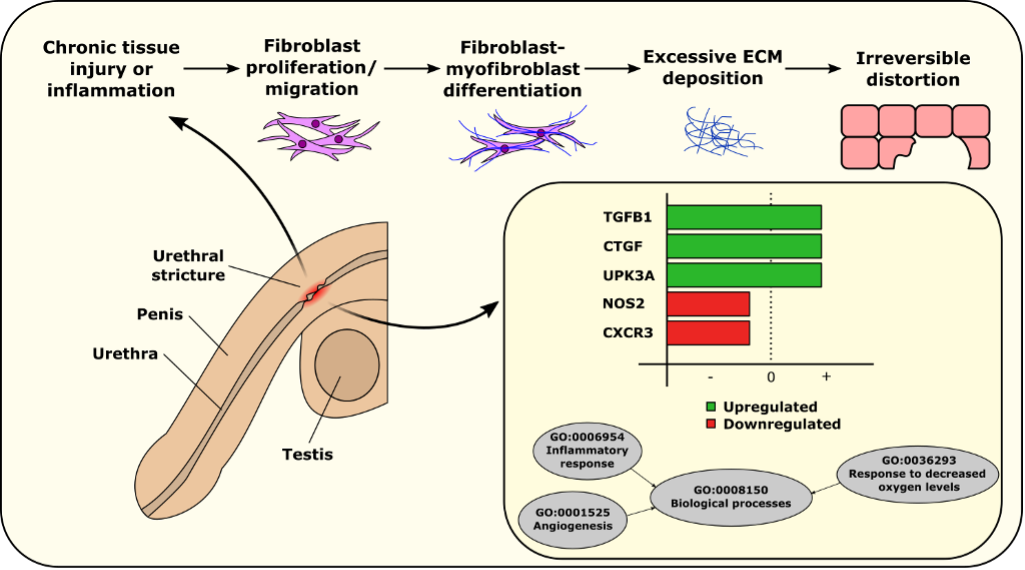
Figure 1. Summary of key genes and pathways in urethral stricture
Our study also identified significant pathways associated with these gene expression changes, particularly highlighting the phosphoinositide 3-kinase (PI3 kinase) and transforming growth factor beta 1/suppressor of mothers against decapentaplegic (TGF-β1/SMAD) signaling pathways.
In conclusion, our systematic review shed light on various gene expression variations and underlying biological pathways linked to urethral stricture. These pivotal findings pave the way for further research and may revolutionize urethral stricture treatment and prevention strategies, offering new hope in the field.
Written by: Ilaha Isali,1 Thomas R. Wong,1 Benjamin N. Breyer2
- Department of Urology, University Hospitals, Cleveland Medical Center, Cleveland, OH, USA
- Department of Urology, University of California San Francisco, San Francisco, CA, USA
Read the Abstract


