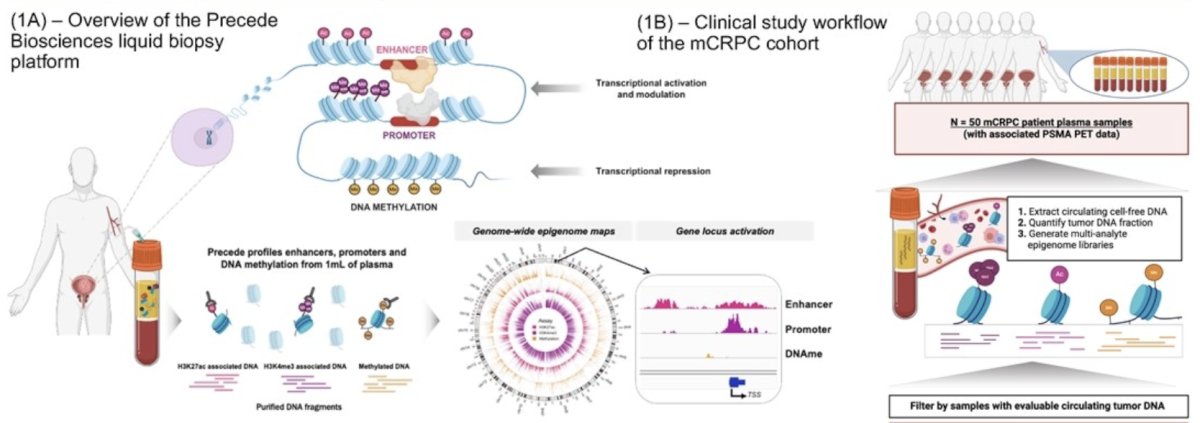
(UroToday.com) The 2024 ESMO annual meeting included a session on prostate cancer, featuring a presentation by Dr. Praful Ravi discussing the determination of tumor PSMA expression in prostate cancer from blood using a novel epigenomic liquid biopsy platform. PSMA, a highly expressed cell surface protein in prostate cancer, has emerged as a pivotal biomarker for both diagnosis and therapeutic targeting in patients with advanced disease. 177Lu-PSMA-617 is an FDA-approved radiopharmaceutical therapy for men with metastatic castration-resistant prostate cancer (mCRPC).1 However, treatment eligibility requires a PSMA PET scan to confirm tumor PSMA expression. With therapeutic strategies in development targeting an array of cell surface proteins, there is an emerging unmet need to quantify tumor drug target expression minimally invasively. Dr. Ravi and colleagues performed comprehensive epigenomic profiling on 1 mL of plasma and demonstrated an accurate minimally invasive readout of tumor PSMA expression in mCRPC.
This study collected plasma at the time of PSMA PET scan in 50 men with mCRPC and analyzed the PET images to quantify total tumor SUVmean, SUVmax, and tumor volume (PSMA-avid disease with voxel SUV>3). The investigators profiled genome-wide epigenomic signals across histone modifications associated with active enhancers, promoters, and DNA methylation on these plasma samples and healthy controls. They also identified genes associated with greater gene-regulatory activation signal in men with mCRPC and evaluated the association of this signal with PSMA PET features. Plasma samples with detectable ctDNA (>= 3%) were used to train a multi-analyte model on FOLH1 (PSMA) epigenomic signal to predict PSMA PET SUVmean. Performance was assessed in a leave-one-out cross-validation schema and in an independent validation cohort. The comprehensive epigenomic platform offers dynamic resolution into target and pathway biology from 1mL of plasma:
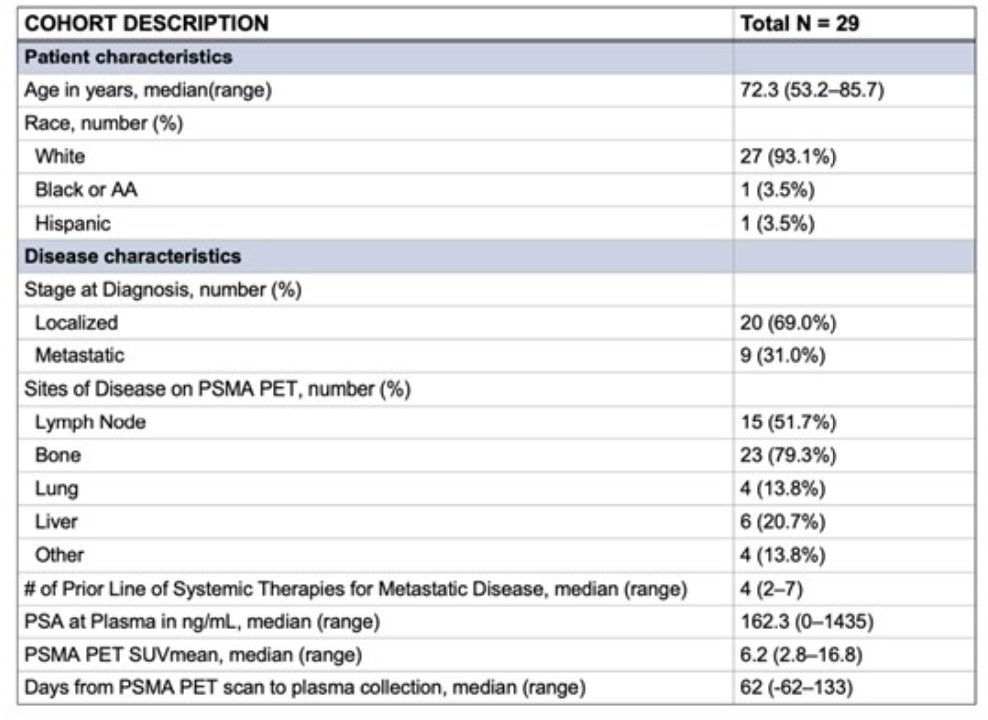
At ESMO 2024, Dr. Ravi presented results for 29 mCRPC patients with the following baseline characteristics:
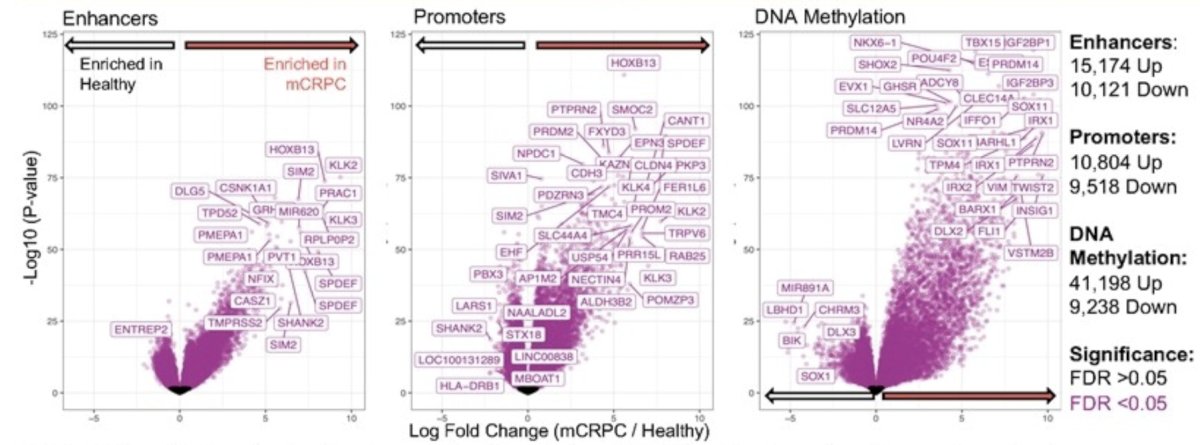
Differential analysis of normalized epigenomic signal between mCRPC patients (n = 29) and healthy male volunteers (n = 51) revealed enrichment of multiple prostate cancer specific signals including HOXB13, KLK2, KLK3, and SPDEF in mCRPC patient plasma. Fold enrichment and significance were calculated using DESeq with Benjamini-Hochberg p-value correction. Points represent individual peaks and are colored by significance (purple = FDR <0.05, Black = FDR > 0.05), and “up”/”down” labels indicate the number of statistically significant loci for enhancers, promoters, and DNA methylation that are upregulated or downregulated in mCRPC patients versus healthy volunteers:
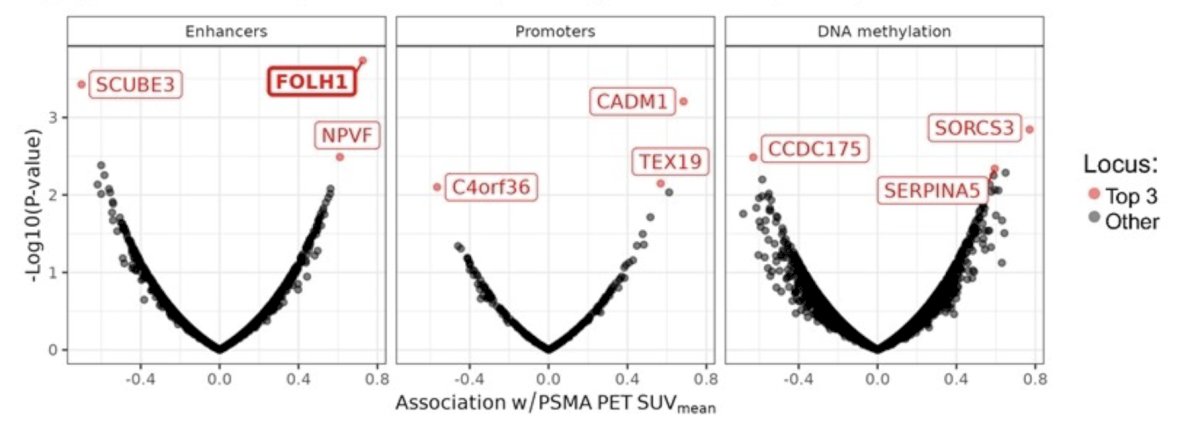
A genome wide analysis correlating epigenomic signal to PSMA PET SUVmean quantifications in mCRPC patients (ctDNA% >= 3, n = 29) identifies enhancer signal at the FOLH1 locus as the top association. For each analyte, robust mCRPC–specific epigenomic features were identified and normalized to reduce technical variability and dependence on ctDNA fraction. Each feature was then tested for its association with PSMA PET SUVmean via linear regression:
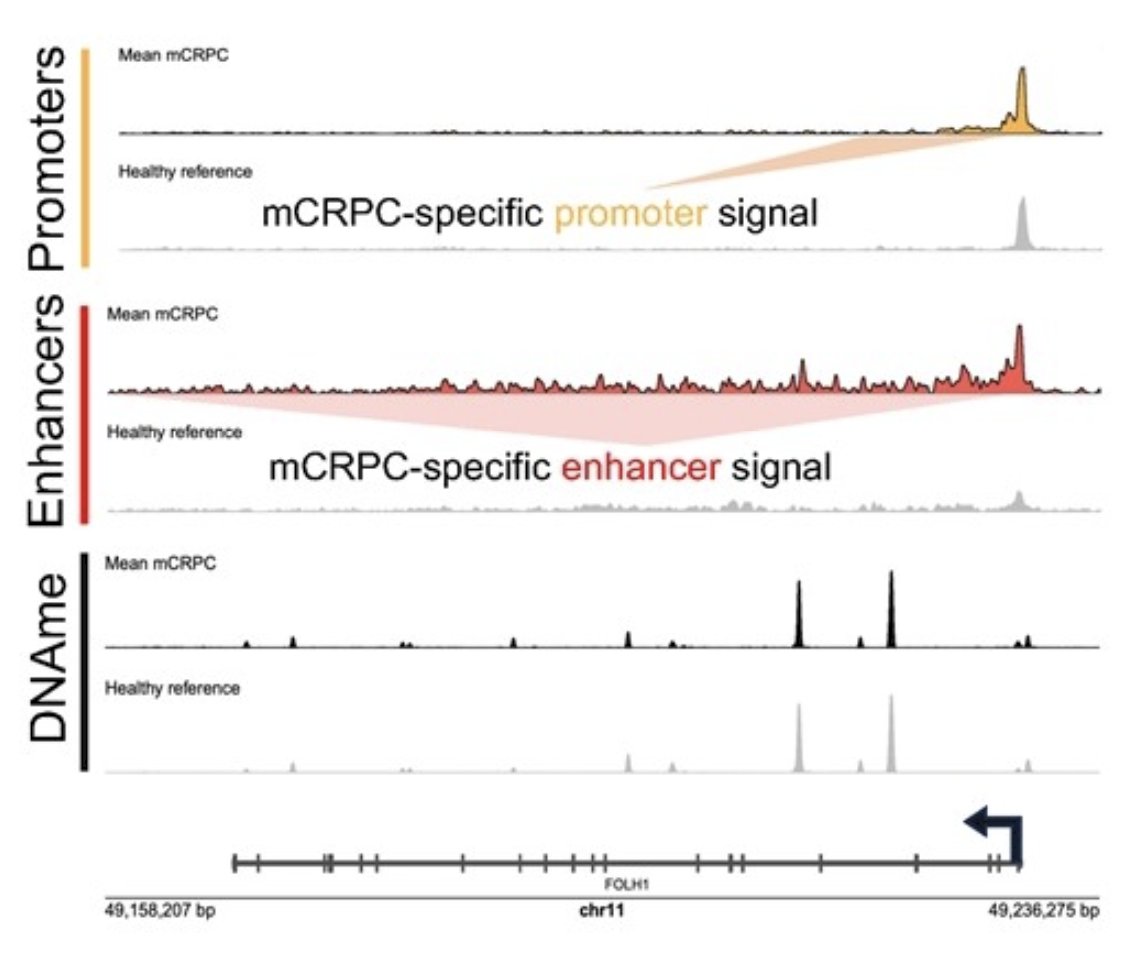
Epigenomic signal at the FOLH1 locus shows increased active promoter and active enhancer marks in mCRPC patients compared to healthy volunteers. Enhancer, promoter, and DNA methylation signals in mCRPC patients were normalized, smoothed, and averaged together with the mean signal from a cohort of male healthy volunteers:
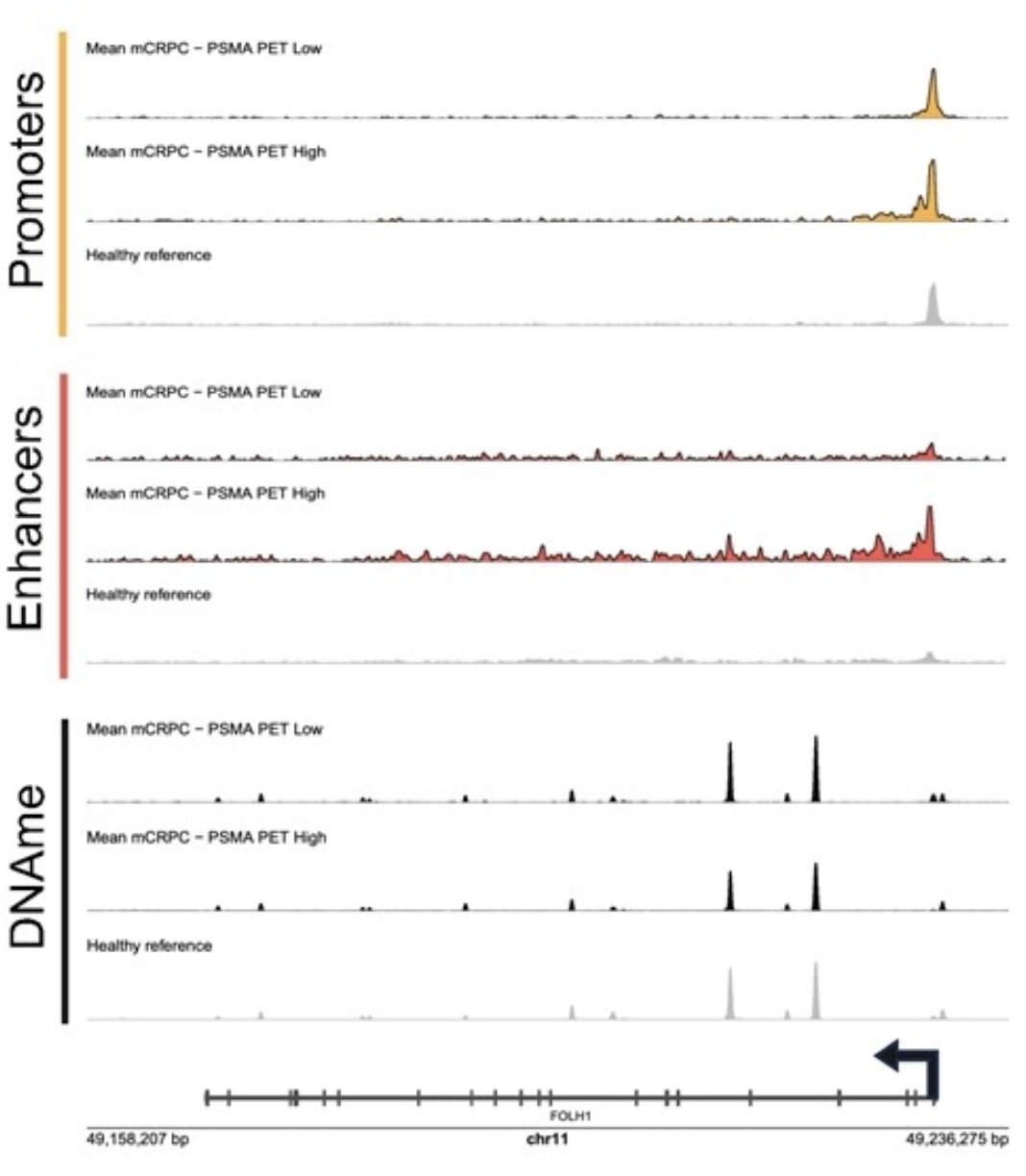
Epigenomic signal at the FOLH1 locus shows increased active promoter and active enhancer marks in mCRPC patients with high PSMA PET SUVmean (>= median, n = 15) compared to mCRPC patients with low PSMA PET SUVmean (< median, n = 14). Enhancer, promoter, and DNA methylation signal in mCRPC patients with either high or low PSMA PET SUVmean was normalized, smoothed, and averaged together (within analyte) with the mean signal from a cohort of male healthy volunteers:
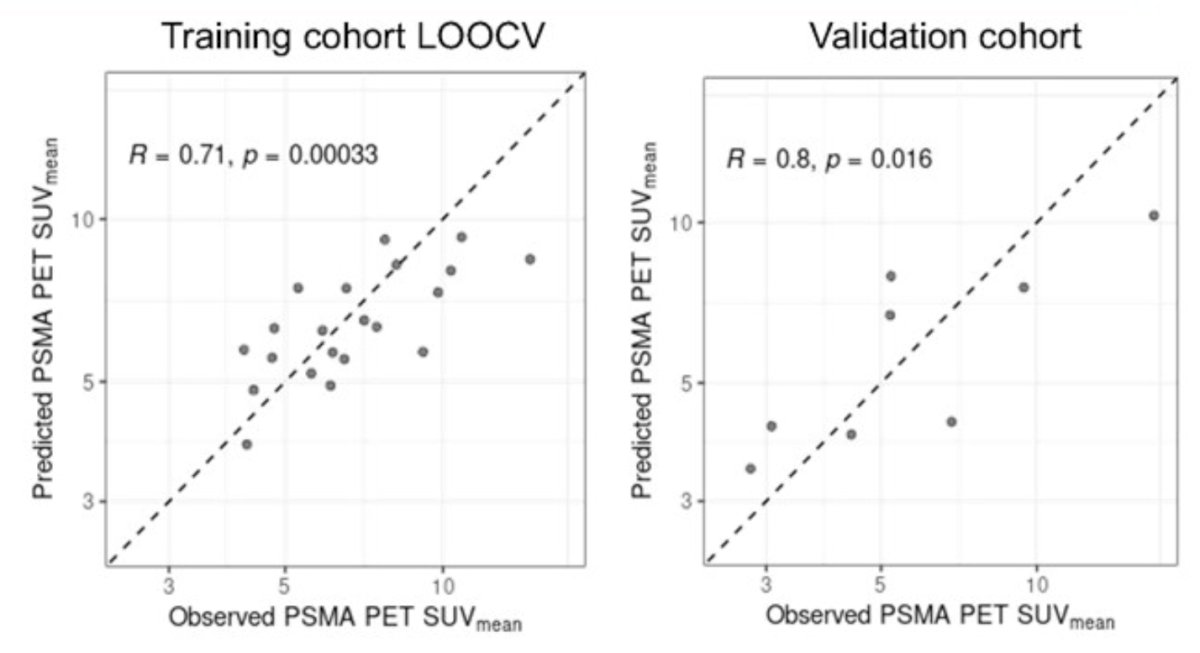
Enhancer and promoter signals at the FOLH1 locus were used to train a machine learning model to predict PSMA PET SUVmean. Samples were first split into training and validation cohorts, which were matched for ctDNA% and PSMA PET SUVmean distributions. For model training, training cohort samples (ctDNA% >= 3) were used to identify robust, mCRPC-specific enhancer/promoter regions at the FOLH1 locus. Signals at these regions were then used to train a model to predict the corresponding PSMA PET SUVmean quantifications. Performance was assessed via Pearson correlation in both a leave it out cross validation setting within the training cohort, as well as the held out validation cohort using a final model trained on all data from the training cohort:
Dr. Ravi concluded his presentation by discussing the determination of tumor PSMA expression in prostate cancer from blood using a novel epigenomic liquid biopsy platform with the following take-home points:
- This retrospective study profiled 50 patients with mCRPC who were treated with 177Lu-PSMA-617 based on PSMA PET evaluation
- Evaluating PSMA levels from blood is crucial for optimizing patient selection and treatment efficacy in the growing landscape of PSMA-targeted therapies. The current analysis suggests that comprehensive epigenomic cfDNA profiling provides an accurate surrogate of tumor PSMA expression in men with mCRPC
- With myriad drugs in development targeting a wide array of cancer-enriched cell surface proteins, these results highlight the potential of epigenomic cfDNA profiling as a real-time, expedient, and non-invasive readout of tumor drug target expression
- Future work will be aimed at increasing the accuracy and sensitivity of the PSMA predictive model, as well as examining the association of these plasma epigenomic signal-based PSMA PET predictions with response to 177Lu-PSMA-617 and other PSMA-targeted therapies
Presented by: Praful Ravi, MB, BChir, MRCP, Dana Farber Cancer Center, Boston, MA
Written by: Zachary Klaassen, MD, MSc – Urologic Oncologist, Associate Professor of Urology, Georgia Cancer Center, Wellstar MCG Health, @zklaassen_md on Twitter during the 2024 European Society of Medical Oncology (ESMO) Annual Meeting, Barcelona, Spain, Fri, Sept 13 – Tues, Sept 17, 2024.
References:


