(UroToday.com) FGFR3 has been one of the most heavily studied and scrutinized proteins along the continuum of urothelial carcinogenesis. Mutations in receptor tyrosine kinases have been described in up to 30% of muscle-invasive bladder cancer (MIBC), with FGFR3 being implicated in more than 50% of these cases. Rates of FGFR3 mutations vary considerably by stage and grade, with mutations in non-muscle-invasive bladder cancer (NMIBC) ranging from 60-70%.1
Dr. Francois Radvanyi provides historical context on the immense preclinical studies informing our knowledge of FGFR3 signaling today. FGFR3 mutational burden has been characterized in dysplastic disease states in nonurothelial tissues, including bone (skeletal dysplasias) and skin (seborrheic keratoses and cutaneous malignancy; Figure 1).2
Figure 1. Incidence and characterization of FGFR3 SNPs in bladder cancer and skeletal dysplasias.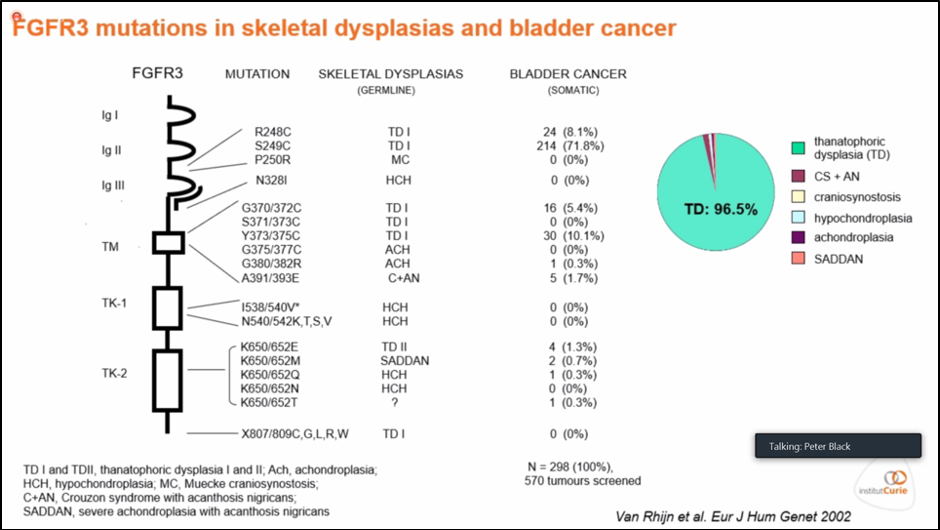
FGFR3 appears to have both oncogenic and tumor suppressor roles, as its expression has a reverse relationship with stage and grade. High mutational burdens are observed in TaLG disease. However, FGFR3 does not appear to be the first activating mutation during tumorigenesis, as these mutations are commonly temporally preceded by homozygous loss of CDK2A.3
Activating somatic SNP S249C is the most commonly observed FGFR3 mutation in urothelial cancers.2 Conditional mutational mouse models have been generated with both Keratin 5 and Uroplakin II promoters (Figures 2-3). Conditional expression utilizing the K5 promoter induced benign proliferations in the skin, while the UPII promoter induced urothelial dysplasia with low-grade carcinomas in an allele-dependent fashion, with >50% homozygous male mice growing bladder tumors.
Figure 2. Conditional FGFR3 S249C expression in mouse model driven by Keratin 5 promoter.
Figure 3. Conditional FGFR3 S249C expression in mouse model driven by Uroplakin II promoter.
APOBEC (apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like) is an mRNA editing enzyme with a proposed homeostatic role of increasing protein diversity. There are at least 10 functional isoenzymes in this family, and it is estimated that more than 50% of point mutations in bladder cancer are due to APOBEC. The only FGFR3 mutation known to associate with an APOBEC motif is S259C, with this SNP associated with a high APOBEC mutational signature compared to other mutated tumors (Figure 4).4
Figure 4. Association with FGFR3 S249C mutations and APOBEC mutagenesis.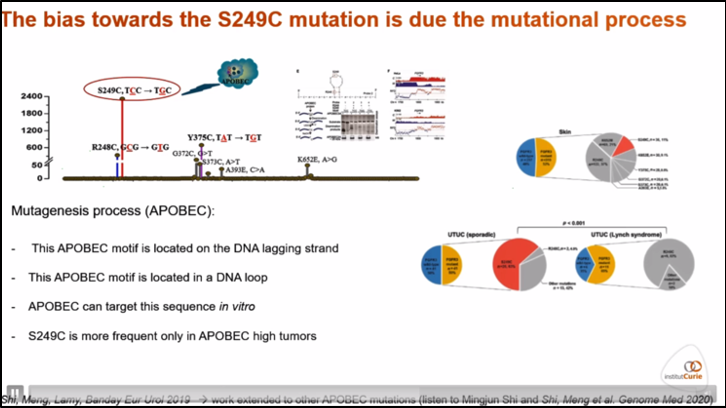
Dr. Radvanyi and his colleagues then profiled the downstream signaling effects of PD173074, a ubiquitous RTK inhibitor with potent anti-FGFR3 activity, in both in vitro and in vivo models with induced variable expression of FGFR3. A positive feedback loop was identified between FGFR3 and MYC, a proto-oncogene (Figure 5). Inhibition of FGFR3 downregulated both its primary target and, secondarily, MYC expression, with resultant anti-tumor efficacy.5
Figure 5. Positive feedback loop between FGFR3 and MYC.
Dr. Radvanyi concluded with the contextualization of FGFR3 mutational signatures within molecular bladder cancer subtypes. The differentiated luminal papillary subtypes are heavily enriched in FGFR3 mutations relative to basal/squamous and NE-like subtypes. These data provide a rationale for the use of recently approved FGFR-inhibitors, including erdafitinib approved for the use of advanced urothelial cancer patients with FGFR2/3 mutations, in earlier disease states along the stage and grade continuum. Further data is necessary to understand the role of these inhibitors in non-luminal papillary subtypes, or in those patients within this subtype without identifiable FGFR mutations.
Presented by: Francois Radvanyi, MD, Professor, Institut Curie, Paris, France.
Written by: Dr. Patrick Hensley, Urologic Oncology Fellow at MD Anderson Cancer Center, Twitter: @pjhensley11, with Ashish Kamat, MD, MBBS, President of The International Bladder Cancer Network (IBCN), The International Bladder Cancer Group (IBCG), and Professor of the Department of Urology, Division of Surgery, The University of Texas MD Anderson Cancer Center, Houston, Texas, Twitter: @UroDocAsh, at the International Bladder Cancer Network (IBCN) Annual Meeting, #IBCN2020, October 17, 2020.
References:
2. van Rhijn BW, van Tilborg AA, Lurkin I, Bonaventure J, de Vries A, Thiery JP, van der Kwast TH, Zwarthoff EC, Radvanyi F. Novel fibroblast growth factor receptor 3 (FGFR3) mutations in bladder cancer previously identified in non-lethal skeletal disorders. Eur J Hum Genet. 2002 Dec;10(12):819-24. doi: 10.1038/sj.ejhg.5200883. PMID: 12461689.
3. di Martino E, L'Hôte CG, Kennedy W, Tomlinson DC, Knowles MA. Mutant fibroblast growth factor receptor 3 induces intracellular signaling and cellular transformation in a cell type- and mutation-specific manner. Oncogene. 2009 Dec 3;28(48):4306-16. doi: 10.1038/onc.2009.280. Epub 2009 Sep 14. PMID: 19749790; PMCID: PMC2789045.


