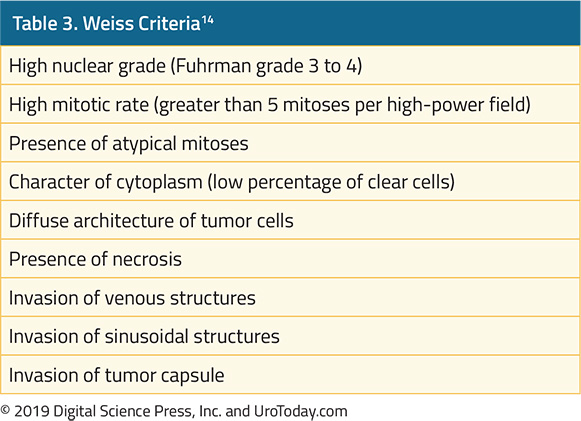
Prostate Cancer and Tumor Markers
Blood
PSAPSA is part of the kallikrein gene family located on chromosome 19 and functions as a serine protease, predominantly produced by prostate luminal cells. PSA in the serum is typically bound to proteins (~80% of PSA; complexed) or unbound (free PSA). The production of PSA is androgen dependent4 and in the absence of cancer varies with age,5 race,6, 7 and prostate volume.8 African-American men without prostate cancer have a higher PSA level compared to similar Caucasian men when assessed on a volume-to-volume ratio.9 Additionally, many studies have suggested that PSA in men with higher body mass index (BMI) have lower PSAs, a concept referred to as “hemodilution”:10 a greater plasma volume leading to lower hematocrit and PSA. Recent studies have provided further support for the hemodilution theory, in that only a fraction of lower PSA values in obese men are attributed to testosterone and dihydrotestosterone levels, with the remaining lower PSA explained presumably by hemodilution.11 The greatest contributor to elevated PSA is prostatic diseases, namely prostatitis, BPH and prostate cancer. Without question, the decrease in specificity associated with PSA and prostate cancer is an elevated PSA in men with prostatitis and/or BPH.
Free PSA (fPSA)
fPSA is PSA that is enzymatically inactive and non-complexed, making up 4-45% of total PSA;12 men with PSA from prostate cancer cells have a lower percentage of total PSA that is free, compared to those without prostate cancer.13 fPSA has FDA approval for men with a negative digital rectal examination (DRE) and total PSA level of 4-10 ng/mL, largely on the basis of a prospective study of men demonstrating a %fPSA (fPSA/total PSA) cutoff of 25% detecting 95% of prostate cancers, while avoiding 20% of biopsies.14 A generally acceptable cut-point ranges from 15-25%. Twenty years later, %fPSA is still used for clinically appropriate men, most commonly used in those with an elevated PSA and a negative prostate biopsy. In these men, studies have reported a 5% cancer under-detection rate and 21% cutoff for repeating prostate biopsy.15
Kallikreins
PSA, also known as human kallikrein 3 (hK3), is the most famous of the kallikreins, however, there are other kallikreins that have recently been explored as prostate cancer tumor markers. hK2 shares 80% amino acid homology with PSA, however, is weakly expressed in benign tissue and intensely expressed in prostate cancer tissue.16 Low-grade disease generally has low expression of hK2, whereas aggressive disease has high levels of expression.16 Recently, the hK2 kallikrein has been incorporated into a panel of kallikrein markers (total PSA, free PSA, intact PSA, and hK2, along with clinical information), commercially available as the 4KScore Test, used for calculating a patient’s percent risk for aggressive prostate cancer. First described in 2008, Vickers et al.17 tested the utility of the kallikrein panel in 740 men in the Swedish arm of the ERSPC screening trial. They found that adding free and intact PSA with hK2 to total PSA improved the clinical area under the curve (AUC) from 0.72 to 0.84. When the authors applied a 20% risk of prostate cancer as the threshold for biopsy, 424 (57%) of biopsies would have been avoided, missing 31 of 152 low-grade and 3 of 40 high-grade cancers.17 Since this study a decade ago, many studies have validated these findings, including among 6,129 men participating in the ProtecT study:18 the AUC for the four kallikreins was 0.719 (95%CI 0.704-0.734) vs 0.634 (95%CI 0.617-0.651, p<0.001) for PSA and age alone for any-grade cancer, and 0.820 (95%CI 0.802-0.838) vs 0.738 (95%CI 0.716-0.761) for high-grade prostate cancer.
Prostate Health Index (phi)
The phi test combines total, free and [-2]proPSA into a single score for improving the accuracy of prostate cancer detection. In the seminal study leading to FDA approval, Catalona et al.19 assessed phi scores among 892 patients without prostate cancer and a PSA between 2-10 ng/mL. They found that an increasing phi score was associated with a 4.7-fold increased risk of prostate cancer and a 1.6-fold increased risk of Gleason score ≥ 4+3 disease at prostate biopsy. Furthermore, the phi score AUC exceeded that of %fPSA (0.72 vs 0.67) to discriminate high vs low-grade disease or negative biopsy. In a subsequent study, Loeb et al.20 confirmed the phi score’s ability to outperform total, free and [-2]proPSA for identifying clinically significant prostate cancer.
Urine
Prostate Cancer Antigen 3 (PCA3)PCA3 is a long noncoding RNA shed into the urine that is not expressed outside the prostate and is associated with much higher expression in malignant than benign prostate tissue.21 Prior to collecting urine for a PCA3 test, a “rigorous” DRE is performed in order to enhance the sensitivity of the test. The commercial PCA3 score is reported as a ratio of urine PCA3 mRNA to urine PSA mRNA x 1000. The optimal cutoff is still debated, however in a contemporary comparative effectiveness review, Bradley et al.22 showed that a PCA3 threshold of 25 resulted in a sensitivity of 74% and specificity of 57% for a positive biopsy. This threshold led to FDA approval of the PCA3 test in 2012 among men with a prior negative prostate biopsy.
Since then, several groups have reported results of PCA3 in biopsy naïve men. In a retrospective review of 3,073 men undergoing initial biopsy, Chevli et al.23 found that the mean PCA3 was 27.2 for those without, and 52.5 for patients with prostate cancer. Prostate cancer was identified in 1,341 (43.6%) men; on multivariable analysis, PCA3 was associated with any (OR 3.0, 95%CI 2.5-3.6) and high-grade (OR 2.4, 95%CI 1.9-3.1) prostate cancer after adjusting for clinicopathologic variables. Furthermore, PCA3 outperformed PSA in the prediction of prostate cancer (AUC 0.697 vs 0.599, p<0.01) but did not for high-grade disease (AUC 0.682 vs 0.679, p=0.702).23
microRNAs (miRNAs)
miRNAs are small, noncoding single-stranded RNAs involved in the regulation of mRNA. Due to their short sequence (typically 19-22 nucleotides), miRNAs are highly stable in most body fluids (including urine) as they are resistant to RNase degradation.24 Several miRNAs have been implicated as potential biomarkers in prostate cancer diagnosis and management, including miRNA-141, miRNA-375, miRNA-221, miRNA-21, miRNA-182 and miRNA-187.25, 26 miR-187 detected in urine has been suggested as a candidate for improving the predictive value for a positive biopsy; a prediction model including serum PSA, urine PCA3, and miR-187 provided 88.6% sensitivity and 50% specificity (AUC 0.711, p = 0.001) for a positive biopsy.26 Ultimately, these miRNAs need to be further validated in terms of their ability to regulate various pathways important for prostate cancer management and their potential role as tumor markers.
Combining Tumor Markers
In an effort to improve the predictive accuracy of a positive biopsy, the last several years have seen a plethora of studies combining biomarkers to not only improve predictive accuracy above that offered by PSA, but also individual, newer biomarkers. As previously mentioned, the decrease in specificity associated with PSA and prostate cancer is secondary to an elevated PSA in men with prostatitis and/or BPH. The “perfect” biomarker (or combination) would delineate prostate cancer (and ultimately high-grade prostate cancer) from other benign entities.Vedder et al.27 assessed the added value of %fPSA, PCA3, and 4KScore Test to the ERSPC prediction models among men in the Dutch arm of the ERSPC screening trial. Prostate cancer was detected in 119 of 708 men – adding %fPSA did not improve the predictive value of the risk calculators, however, the 4KScore discriminated better than PCA3 in univariate models (AUC 0.78 vs. 0.62; p=0.01). In the overall population, there was no statistically significant difference between the multivariable model with PCA3 (AUC 0.73) versus the model with the 4KScore (AUC 0.71; p=0.18). Among 127 men with a previous negative biopsy, Auprich et al.28 compared the performance of total PSA, %fPSA, PSA velocity (PSAV), and PCA3 at first, second and ≥ third repeat biopsy. At first repeat biopsy, PCA3 predicted prostate cancer best (AUC 0.80) compared with total PSA. A second repeat biopsy, %fPSA demonstrated the highest accuracy (AUC 0.82), and again at ≥ third repeat biopsy %fPSA demonstrated the highest accuracy (AUC 0.70).28
This sampling of studies demonstrates that many combinations of biomarkers are being studied in an effort to improve detection of high-grade cancer and decrease the number of unnecessary biopsies. The next generation of biomarker combinations has and will continue to incorporate multi-parametric prostate MRI into predictive algorithms for clinically significant prostate cancer.29-31
Conclusions
For over four decades, research efforts have been directed towards improving the detection of prostate cancer and attempting to build on the predictive accuracy of the first prostate cancer tumor marker, PSA. With the United States Preventative Services Task Force’s 2012 recommendation for the urgent need to identify new screening efforts to better identify indolent versus aggressive disease, the last several years have seen a dramatic increase in prostate cancer biomarker options. As briefly highlighted, biomarker combinations studies have demonstrated improved predictive accuracy of positive biopsies; however, these combinations are far from perfect, are expensive and much work remains to be done. Furthermore, the specific indication (pre-biopsy, post-negative biopsy, active surveillance, etc) and a combination of tumor markers remain to be fully elucidated.Published Date: April 16th, 2019